Abstract
We present novoBreak, a genome-wide local assembly algorithm that discovers somatic and germline structural variation breakpoints in whole-genome sequencing data. novoBreak consistently outperformed existing algorithms on real cancer genome data and on synthetic tumors in the ICGC-TCGA DREAM 8.5 Somatic Mutation Calling Challenge primarily because it more effectively utilized reads spanning breakpoints. novoBreak also demonstrated great sensitivity in identifying short insertions and deletions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Mitelman, F., Johansson, B. & Mertens, F. Nat. Rev. Cancer 7, 233–245 (2007).
Stephens, P.J. et al. Nature 462, 1005–1010 (2009).
Alkan, C., Coe, B.P. & Eichler, E.E. Nat. Rev. Genet. 12, 363–376 (2011).
Alkan, C., Sajjadian, S. & Eichler, E.E. Nat. Methods 8, 61–65 (2011).
Chen, K. et al. Nat. Methods 6, 677–681 (2009).
Abyzov, A., Urban, A.E., Snyder, M. & Gerstein, M. Genome Res. 21, 974–984 (2011).
Ye, K., Schulz, M.H., Long, Q., Apweiler, R. & Ning, Z. Bioinformatics 25, 2865–2871 (2009).
Rausch, T. et al. Bioinformatics 28, i333–i339 (2012).
Chen, K. et al. Genome Res. 24, 310–317 (2014).
Li, Y. et al. Nat. Biotechnol. 29, 723–730 (2011).
Earl, D. et al. Genome Res. 21, 2224–2241 (2011).
Boutros, P.C. et al. Nat. Genet. 46, 318–319 (2014).
Ewing, A.D. et al. Nat. Methods 12, 623–630 (2015).
McKenna, A. et al. Genome Res. 20, 1297–1303 (2010).
Saunders, C.T. et al. Bioinformatics 28, 1811–1817 (2012).
Li, H. Bioinformatics 28, 1838–1844 (2012).
Pleasance, E.D. et al. Nature 463, 191–196 (2010).
Wang, J. et al. Nat. Methods 8, 652–654 (2011).
Zhang, J. et al. Genome Res. 26, 108–118 (2016).
Li, H. Preprint at https://arxiv.org/abs/1303.3997 (2013).
Bloom, B.H. Communications of the ACM 13, 422–426 (1970).
Sedgewick, R. & Wayne, K. Algorithms 4th edn. (Addison-Wesley, 2011).
Warren, R.L., Sutton, G.G., Jones, S.J.M. & Holt, R.A. Bioinformatics 23, 500–501 (2007).
DePristo, M.A. et al. Nat. Genet. 43, 491–498 (2011).
Van der Auwera, G.A. et al. Curr. Protoc. Bioinformatics 11, 43.11.10.1–43.11.10.33 (2013).
Acknowledgements
We thank the ICGC-TCGA DREAM SMC Challenge organizers and participants for providing data and evaluations; and we thank A.K. Eterovic and G.B. Mills for assistance with the experiment and manuscript. This study was supported in part by the National Institutes of Health (grant numbers R01 CA172652 to K.C. and U41 HG007497 to C. Lee, Jackson Lab), the National Cancer Institute Cancer Center Support Grant (P30 CA016672 to R. Depinho, MD Anderson Cancer Center), Andrew Sabin Family Foundation to K.C., and a training fellowship from the Computational Cancer Biology Training Program of the Gulf Coast Consortia (CPRIT grant number RP140113) to Z.C. The results published here are in part based upon data generated by TCGA established by the NCI and NHGRI. Information about TCGA and the investigators and institutions who constitute the TCGA research network can be found at http://cancergenome.nih.gov/.
Author information
Authors and Affiliations
Contributions
Z.C., J.R., and K.C. conceived the algorithm. Z.C. developed the software. Z.C. and K.C. designed and analyzed the experiments. M.G. and J.C. designed and performed the validation experiments. W.Z. designed the scoring statistics. M.G., T.C., X.F., L.D., A.Y.L., and P.B. tested the algorithm and performed additional analyses. K.C. supervised the projects. Z.C. and K.C. wrote the manuscript with input from all authors. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
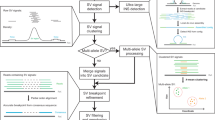
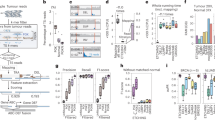
Supplementary Figure 1 Illustration of the fate of the breakpoint spanning read pairs in alignment-based methods versus that in our k-mer targeted assembly method.
The alignment-based approaches underutilize read pairs spanning a breakpoint (top), while our k-mer targeted assembly strategy can fully utilize them (bottom).
Supplementary Figure 2 Illustration of clustering based on novo k-mers and associated read pairs.
At each breakpoint, there are k-1 k-mers covering the breakpoint. If a read fully covers a breakpoint, it must contain several k-mers (< k-1 if there are sequencing errors) covering the breakpoint. On the other hand, there should be several read pairs sharing identical k-mers, given sufficient coverage. Based on this relationship, a union-find algorithm is applied to accomplish the clustering procedure.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2, Supplementary Tables 1, 2 and 6, and Supplementary Notes 1–4. (PDF 674 kb)
Supplementary Table 3
Previously experimentally validated somatic SV breakpoints in COLO-829 cell line. (XLSX 67 kb)
Supplementary Table 4
novoBreak predicted breakpoints in the COLO-829 whole genome sequencing data. (XLSX 47 kb)
Supplementary Table 5
Experimentally validated novel breakpoints found by novoBreak in COLO-829. (XLSX 41 kb)
Supplementary Table 7
All filtered novoBreak calls of the 22 TCGA samples. (XLSX 1169 kb)
Supplementary Table 8
Sensitivity of novoBreak in detecting gene fusions from the whole-genome sequencing data of 22 breast cancer patients from TCGA. (XLSX 48 kb)
Supplementary Software
novoBreak source code and binary distribution. (ZIP 2005 kb)
Rights and permissions
About this article
Cite this article
Chong, Z., Ruan, J., Gao, M. et al. novoBreak: local assembly for breakpoint detection in cancer genomes. Nat Methods 14, 65–67 (2017). https://doi.org/10.1038/nmeth.4084
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.4084
This article is cited by
-
Analysis of Preimplantation and Clinical Outcomes of Two Cases by Oxford Nanopore Sequencing
Reproductive Sciences (2024)
-
Personalized genome assembly for accurate cancer somatic mutation discovery using tumor-normal paired reference samples
Genome Biology (2022)
-
Structural variations in cancer and the 3D genome
Nature Reviews Cancer (2022)
-
Ultrafast prediction of somatic structural variations by filtering out reads matched to pan-genome k-mer sets
Nature Biomedical Engineering (2022)
-
Detecting structural variations with precise breakpoints using low-depth WGS data from a single oxford nanopore MinION flowcell
Scientific Reports (2022)