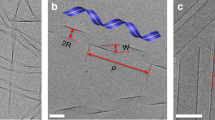
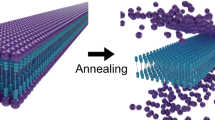
Abstract
A large variety of functional self-assembled supramolecular nanostructures have been reported over recent decades1. The experimental approach to these systems initially focused on the design of molecules with specific interactions that lead to discrete geometric structures1,2,3,4, and more recently on the kinetics and mechanistic pathways of self-assembly5,6. However, there remains a major gap in our understanding of the internal conformational dynamics of these systems and of the links between their dynamics and function. Molecular dynamics simulations have yielded information on the molecular fluctuations of supramolecular assemblies5,6,7, yet experimentally it has been difficult to obtain analogous data with subnanometre spatial resolution. Using site-directed spin labelling and electron paramagnetic resonance spectroscopy, we measured the conformational dynamics of a self-assembled nanofibre in water through its 6.7 nm cross-section. Our measurements provide unique insight for the design of supramolecular functional materials.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Palmer, L. C. & Stupp, S. I. Molecular self-assembly into one-dimensional nanostructures. Acc. Chem. Res. 41, 1674–1684 (2008).
Hartgerink, J. D., Beniash, E. & Stupp, S. I. Self-assembly and mineralization of peptide-amphiphile nanofibers. Science 294, 1684 (2001).
Whitesides, G. M. & Grzybowski, B. Self-assembly at all scales. Science 295, 2418–2421 (2002).
Hill, J. P. et al. Self-assembled hexa-peri-hexabenzocoronene graphitic nanotube. Science 304, 1481–1483 (2004).
Klein, M. L. & Shinoda, W. Large-scale molecular dynamics simulations of self-assembling systems. Science 321, 798–800 (2008).
Lee, O. S., Stupp, S. I. & Schatz, G. C. Atomistic molecular dynamics simulations of peptide amphiphile self-assembly into cylindrical nanofibers. J. Am. Chem. Soc. 133, 3677–3683 (2011).
Venable, R. M., Zhang, Y., Hardy, B. J. & Pastor, R. W. Molecular dynamics simulations of a lipid bilayer and of hexadecane: An investigation of membrane fluidity. Science 262, 223–226 (1993).
Shaw, D. E. et al. Atomic-level characterization of the structural dynamics of proteins. Science 330, 341–346 (2010).
Buchete, N-V., Tycko, R. & Hummer, G. Molecular dynamics simulations of Alzheimer’s β-amyloid protofilaments. J. Mol. Biol. 353, 804–821 (2005).
McHaourab, H. S., Lietzow, M. A., Hideg, K. & Hubbell, W. L. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry 35, 7692–7704 (1996).
Török, M. et al. Structural and dynamic features of Alzheimer’s Aβ peptide in amyloid fibrils studied by site-directed spin labeling. J. Biol. Chem. 277, 40810–40815 (2002).
Newcomb, C. J. et al. Cell death versus cell survival instructed by supramolecular cohesion of nanostructures. Nature Commun. 5, 3321 (2014).
Gilbert, P. M. et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Sci. Signaling 329, 1078 (2010).
Sur, S. et al. A hybrid nanofiber matrix to control the survival and maturation of brain neurons. Biomaterials 33, 545–555 (2012).
Goldberger, J. E., Berns, E. J., Bitton, R., Newcomb, C. J. & Stupp, S. I. Electrostatic control of bioactivity. Angew. Chem. Int. Ed. 50, 6292–6295 (2011).
Velichko, Y. S., Stupp, S. I. & de la Cruz, M. O. Molecular simulation study of peptide amphiphile self-assembly. J. Phys. Chem. B 112, 2326–2334 (2008).
Paramonov, S. E., Jun, H-W. & Hartgerink, J. D. Self-assembly of peptide-amphiphile nanofibers: The roles of hydrogen bonding and amphiphilic packing. J. Am. Chem. Soc. 128, 7291–7298 (2006).
Paramonov, S. E., Jun, H-W. & Hartgerink, J. D. Self-assembly of peptide-amphiphile nanofibers: The roles of hydrogen bonding and amphiphilic packing. J. Am. Chem. Soc. 128, 7291–7298 (2006).
Toniolo, C., Crisma, M. & Formaggio, F. TOAC, a nitroxide spin-labeled, achiral Cα-tetrasubstituted α-amino acid, is an excellent tool in material science and biochemistry. J. Peptide Sci. 47, 153–158 (1998).
Karim, C. B., Kirby, T. L., Zhang, Z., Nesmelov, Y. & Thomas, D. D. Phospholamban structural dynamics in lipid bilayers probed by a spin label rigidly coupled to the peptide backbone. Proc. Natl Acad. Sci. USA 101, 14437–14442 (2004).
Budil, D. E., Lee, S., Saxena, S. & Freed, J. H. Nonlinear-least-squares analysis of slow-motion EPR spectra in one and two dimensions using a modified Levenberg-Marquardt algorithm. J. Magn. Reson. Ser. A 120, 155–189 (1996).
Levine, H. Thioflavine T interaction with synthetic Alzheimer’s disease β-amyloid peptides: Detection of amyloid aggregation in solution. Protein Sci. 2, 404–410 (1993).
Acknowledgements
This work was supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences and Engineering Division of the US Department of Energy under Award # DE-FG02-00ER45810. J.B.M. acknowledges a postdoctoral fellowship through the National Institutes of Health National Research Service Award # 1F32AR06195501. J.H.O. acknowledges an IBNAM-Baxter early career award. EPR experiments were carried out at The Medical College of Wisconsin, National Biomedical EPR Center, supported by National Institutes of Health Grant P41 EB001980, and also at Northwestern University under support of the National Heart, Lung and Blood Institute of the National Institutes of Health (NIH-HL-13531). X-ray diffraction experiments were carried out at beamline 5ID-D of the Advanced Photon Source at Argonne National Laboratory. Use of the Advanced Photon Source at Argonne National Laboratory was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. This work also made use of Northwestern University’s Biological Imaging Facility (BIF) for electron microscopy and the Integrated Molecular Structure Education and Research Center (IMSERC) for mass spectrometry and FTIR spectroscopy. The authors acknowledge S. Han for helpful discussions and M. Seniw for illustrations.
Author information
Authors and Affiliations
Contributions
J.H.O. designed and performed experiments, and analysed data. C.J.N. synthesized chemicals, performed experiments and analysed data. J.B.M. synthesized chemicals. L.C.P. assisted in data analysis and writing. P.E.D. assisted in performing experiments. L.C.P., B.M.H., S.I.S. and C.J.N. provided intellectual input. J.H.O. and S.I.S. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1275 kb)
Rights and permissions
About this article
Cite this article
Ortony, J., Newcomb, C., Matson, J. et al. Internal dynamics of a supramolecular nanofibre. Nature Mater 13, 812–816 (2014). https://doi.org/10.1038/nmat3979
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmat3979
This article is cited by
-
Mechanistic insights of evaporation-induced actuation in supramolecular crystals
Nature Materials (2021)
-
Domain-selective thermal decomposition within supramolecular nanoribbons
Nature Communications (2021)
-
Delivery of local anaesthetics by a self-assembled supramolecular system mimicking their interactions with a sodium channel
Nature Biomedical Engineering (2021)
-
Self-assembly of aramid amphiphiles into ultra-stable nanoribbons and aligned nanoribbon threads
Nature Nanotechnology (2021)
-
Supramolecular fibrillation of peptide amphiphiles induces environmental responses in aqueous droplets
Nature Communications (2021)