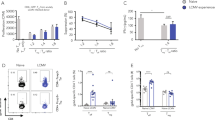
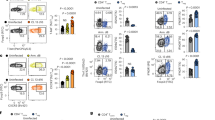
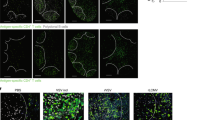
Abstract
Memory CD8+ T cells are critical for host defense upon reexposure to intracellular pathogens. We found that interleukin 10 (IL-10) derived from CD4+ regulatory T cells (Treg cells) was necessary for the maturation of memory CD8+ T cells following acute infection with lymphocytic choriomeningitis virus (LCMV). Treg cell–derived IL-10 was most important during the resolution phase, calming inflammation and the activation state of dendritic cells. Adoptive transfer of IL-10-sufficient Treg cells during the resolution phase 'restored' the maturation of memory CD8+ T cells in IL-10-deficient mice. Our data indicate that Treg cell–derived IL-10 is needed to insulate CD8+ T cells from inflammatory signals, and reveal that the resolution phase of infection is a critical period that influences the quality and function of developing memory CD8+ T cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Ahmed, R. & Gray, D. Immunological memory and protective immunity: understanding their relation. Science 272, 54–60 (1996).
Korber, B.T., Letvin, N.L. & Haynes, B.F. T-cell vaccine strategies for human immunodeficiency virus, the virus with a thousand faces. J. Virol. 83, 8300–8314 (2009).
Kaech, S.M. & Ahmed, R. Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naïve cells. Nat. Immunol. 2, 415–422 (2001).
Kaech, S.M. & Cui, W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12, 749–761 (2012).
Cui, W., Liu, Y., Weinstein, J.S., Craft, J. & Kaech, S.M. An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity 35, 792–805 (2011).
Rutishauser, R.L. et al. Transcriptional repressor Blimp-1 promotes CD8+ T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity 31, 296–308 (2009).
Kim, M.V., Ouyang, W., Liao, W., Zhang, M.Q. & Li, M.O. The transcription factor Foxo1 controls central-memory CD8+ T cell responses to infection. Immunity 39, 286–297 (2013).
Joshi, N.S. et al. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity 27, 281–295 (2007).
Yang, C.Y. et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat. Immunol. 12, 1221–1229 (2011).
Zhou, X. et al. Differentiation and persistence of memory CD8+ T cells depend on T cell factor 1. Immunity 33, 229–240 (2010).
Araki, K. et al. mTOR regulates memory CD8 T-cell differentiation. Nature 460, 108–112 (2009).
Rao, R.R., Li, Q., Gubbels Bupp, M.R. & Shrikant, P.A. Transcription factor Foxo1 represses T-bet-mediated effector functions and promotes memory CD8+ T cell differentiation. Immunity 36, 374–387 (2012).
Haring, J.S. & Harty, J.T. Interleukin-18-related genes are induced during the contraction phase but do not play major roles in regulating the dynamics or function of the T-cell response to Listeria monocytogenes infection. Infect. Immun. 77, 1894–1903 (2009).
Stelekati, E. et al. Bystander chronic infection negatively impacts development of CD8+ T cell memory. Immunity 40, 801–813 (2014).
Sun, J.C., Williams, M.A. & Bevan, M.J. CD4+ T cells are required for the maintenance, not programming, of memory CD8+ T cells after acute infection. Nat. Immunol. 5, 927–933 (2004).
Foulds, K.E., Rotte, M.J. & Seder, R.A. IL-10 is required for optimal CD8 T cell memory following Listeria monocytogenes infection. J. Immunol. 177, 2565–2574 (2006).
Vignali, D.A.A., Collison, L.W. & Workman, C.J. How regulatory T cells work. Nat. Rev. Immunol. 8, 523–532 (2008).
Suvas, S., Kumaraguru, U., Pack, C.D., Lee, S. & Rouse, B.T. CD4+CD25+ T cells regulate virus-specific primary and memory CD8+ T cell responses. J. Exp. Med. 198, 889–901 (2003).
Pace, L. et al. Regulatory T cells increase the avidity of primary CD8+ T cell responses and promote memory. Science 338, 532–536 (2012).
de Goër de Herve, M.G., Jaafoura, S., Vallée, M. & Taoufik, Y. FoxP3+ regulatory CD4 T cells control the generation of functional CD8 memory. Nat. Commun. 3, 986 (2012).
Graham, J.B., Da Costa, A. & Lund, J.M. Regulatory T cells shape the resident memory T cell response to virus infection in the tissues. J. Immunol. 192, 683–690 (2014).
Kühn, R., Löhler, J., Rennick, D., Rajewsky, K. & Müller, W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75, 263–274 (1993).
Maharshak, N. et al. Altered enteric microbiota ecology in interleukin 10-deficient mice during development and progression of intestinal inflammation. Gut Microbes 4, 316–324 (2013).
Stelekati, E. & Wherry, E.J. Chronic bystander infections and immunity to unrelated antigens. Cell Host Microbe 12, 458–469 (2012).
Ouyang, W., Rutz, S., Crellin, N.K., Valdez, P.A. & Hymowitz, S.G. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu. Rev. Immunol. 29, 71–109 (2011).
Cui, W., Joshi, N.S., Jiang, A. & Kaech, S.M. Effects of Signal 3 during CD8 T cell priming: Bystander production of IL-12 enhances effector T cell expansion but promotes terminal differentiation. Vaccine 27, 2177–2187 (2009).
Parish, I.A. et al. Chronic viral infection promotes sustained Th1-derived immunoregulatory IL-10 via BLIMP-1. J. Clin. Invest. 124, 3455–3468 (2014).
Brooks, D.G. et al. Interleukin-10 determines viral clearance or persistence in vivo. Nat. Med. 12, 1301–1309 (2006).
Maynard, C.L. et al. Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3− precursor cells in the absence of interleukin 10. Nat. Immunol. 8, 931–941 (2007).
Srivastava, S., Koch, M.A., Pepper, M. & Campbell, D.J. Type I interferons directly inhibit regulatory T cells to allow optimal antiviral T cell responses during acute LCMV infection. J. Exp. Med. 211, 961–974 (2014).
Cheng, G. et al. IL-2 receptor signaling is essential for the development of Klrg1+ terminally differentiated T regulatory cells. J. Immunol. 189, 1780–1791 (2012).
Steinman, R.M., Pack, M. & Inaba, K. Dendritic cells in the T-cell areas of lymphoid organs. Immunol. Rev. 156, 25–37 (1997).
Jung, Y.W., Rutishauser, R.L., Joshi, N.S., Haberman, A.M. & Kaech, S.M. Differential localization of effector and memory CD8 T cell subsets in lymphoid organs during acute viral infection. J. Immunol. 185, 5315–5325 (2010).
Rubtsov, Y.P. et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558 (2008).
Kaech, S.M., Hemby, S., Kersh, E. & Ahmed, R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell 111, 837–851 (2002).
Luckey, C.J. et al. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 103, 3304–3309 (2006).
Agarwal, P. et al. Gene regulation and chromatin remodeling by IL-12 and type I IFN in programming for CD8 T cell effector function and memory. J. Immunol. 183, 1695–1704 (2009).
Amit, I. et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science 326, 257–263 (2009).
Girard-Madoux, M.J.H., Kel, J.M., Reizis, B. & Clausen, B.E. IL-10 controls dendritic cell-induced T-cell reactivation in the skin to limit contact hypersensitivity. J. Allergy Clin. Immunol. 129, 143–150.e1–10 (2012).
Kim, J.M., Rasmussen, J.P. & Rudensky, A.Y. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 8, 191–197 (2007).
Onishi, Y., Fehervari, Z., Yamaguchi, T. & Sakaguchi, S. Foxp3+ natural regulatory T cells preferentially form aggregates on dendritic cells in vitro and actively inhibit their maturation. Proc. Natl. Acad. Sci. USA 105, 10113–10118 (2008).
Borsellino, G. et al. Expression of ectonucleotidase CD39 by Foxp3+ Treg cells: hydrolysis of extracellular ATP and immune suppression. Blood 110, 1225–1232 (2007).
Fallarino, F. et al. Modulation of tryptophan catabolism by regulatory T cells. Nat. Immunol. 4, 1206–1212 (2003).
Hochrein, H. et al. Differential production of IL-12, IFN-α, and IFN-γ by mouse dendritic cell subsets. J. Immunol. 166, 5448–5455 (2001).
Moser, E.K., Hufford, M.M. & Braciale, T.J. Late engagement of CD86 after influenza virus clearance promotes recovery in a FoxP3+ regulatory T cell dependent manner. PLoS Pathog. 10, e1004315 (2014).
Liston, A. & Gray, D.H.D. Homeostatic control of regulatory T cell diversity. Nat. Rev. Immunol. 14, 154–165 (2014).
Zeiser, R. et al. Differential impact of mammalian target of rapamycin inhibition on CD4+CD25+Foxp3+ regulatory T cells compared with conventional CD4+ T cells. Blood 111, 453–462 (2008).
Keating, R. et al. The kinase mTOR modulates the antibody response to provide cross-protective immunity to lethal infection with influenza virus. Nat. Immunol. 14, 1266–1276 (2013).
Cui, W. et al. TLR4 ligands lipopolysaccharide and monophosphoryl lipid a differentially regulate effector and memory CD8+ T Cell differentiation. J. Immunol. 192, 4221–4232 (2014).
Best, J.A. et al. Transcriptional insights into the CD8+ T cell response to infection and memory T cell formation. Nat. Immunol. 14, 404–412 (2013).
Kühn, R., Löhler, J., Rennick, D., Rajewsky, K. & Müller, W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75, 263–274 (1993).
Maynard, C.L. et al. Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3− precursor cells in the absence of interleukin 10. Nat. Immunol. 8, 931–941 (2007).
Roers, A. et al. T cell-specific inactivation of the interleukin 10 gene in mice results in enhanced T cell responses but normal innate responses to lipopolysaccharide or skin irritation. J. Exp. Med. 200, 1289–1297 (2004).
Kaech, S.M. et al. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4, 1191–1198 (2003).
Kim, J.M., Rasmussen, J.P. & Rudensky, A.Y. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 8, 191–197 (2007).
Joshi, N.S. et al. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity 27, 281–295 (2007).
Teijaro, J.R. et al. Cutting edge: Tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J. Immunol. 187, 5510–5514 (2011).
Anders, S. et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 8, 1765–1786 (2013).
Love, M.I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Acknowledgements
We thank J.M.M. den Haan (UV University Medical Center, Amsterdam) for mAb ES5-2A5; and all members of the Kaech and Craft laboratories for discussions and critical reading of the manuscript. Supported by the US National Institutes of Health (RO1AI066232 and R01AI074699 to S.M.K.; R01AR40072, P30AR053495 and R21AR063942 to J.C.; T32AI07019 and F31AG07777 to B.J.L.; and T32GM07205 to S.M.G.) and the Howard Hughes Medical Institute (S.M.K., B.J.L., R.A.F. and T.G.).
Author information
Authors and Affiliations
Contributions
B.J.L., J.C., S.M.K. conceived of and designed the experiments, analyzed the data and wrote the manuscript; and B.J.L., W.C., R.A.A., S.M.G., T.G., Y.L., Y.K., S.H.K. and R.A.F. performed the experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 IL-10 is required for optimal maturation of memory CD8+ NP396+ T cells.
(a) Analysis of the virus-specific CD8+ T cell response 60 days post acute LCMV Armstrong infection. The percentages and numbers of GP33+ T cells along with the (b) representative plots and analysis of the NP396+ T cell response in the spleen of wild type and Il10–/– mice are shown. Percentages and numbers of NP396+ T cells, along with the percentages of CD127+KLRG1-, CD127-KLRG1+, and CD62L+KLRG1- cells in the NP396+ T cell population are shown. (c) Mice were infected with acute LCMV and treated with αIL-10 between days 0-30, 0-8, 8-30, 15-30 or mock injected with PBS. Mice were sacrificed at day 30 and the percentage and numbers of GP33+ T cells determined. Statistical analyses were performed using the unpaired two-tailed Student’s t-test. (*, p < 0.01; **, p < 0.001). Data are from one experiment representative of 5 experiments (a, b) or 3 experiments (c) with at least 4 mice per group carried out 45-60 days (a, b) or 30 days (c) following LCMV Armstrong infection (mean and s.e.m).
Supplementary Figure 2 IL-10 is required in a CD8+ T cell–extrinsic and CD8+ T cell–intrinsic manner to allow the maturation and survival of memory CD8+ T cells.
50,000 Il10raf/f or Il10raf/fCd4-Cre P14 GP33+ cells were transferred into congenically-mismatched mice one day prior to infection with acute LCMV infection. Analysis of the P14 GP33+ T cell response was carried out 30 days p.i. (a) Representative plots and analysis of the P14 GP33+ T cell response. Percentages and numbers of P14 GP33+ T cells, along with the percentages of CD127+KLRG1-, CD127-KLRG1+, and CD62L+KLRG1- cells in the P14 GP33+ T cell population are shown. (b) Representative histograms of GzmB and Tcf1 expression in Il10raf/f (black) or Il10raf/fCd4-Cre (gray filled) P14 GP33+ cells. Statistical analyses were performed using the unpaired two-tailed Student’s t-test. (*, p < 0.05; **, p < 0.01). Data are from one experiment representative of 2 experiments with 4 mice per group carried out 30 days following LCMV Armstrong infection (mean and s.e.m).
Supplementary Figure 3 CD4+ Treg cells continue to produce IL-10 during the resolution phase of infection.
(a) Analysis of the GP33+ T cell response 60 days post acute LCMV infection in Il10f/f, Il10f/fCd4-Cre, Il10f/fLyz2-Cre, and Il10f/fCd11c-Cre mice. Percentages and numbers of GP33+ T cells were determined. (b) Analysis of IL-10 production by T cells following acute LCMV infection in IL-10 reporter (10BiT Thy1.1 mice). Percentages and numbers of IL-10-Thy1.1 reporter-positive cells at multiple time points post LCMV infection are shown. (c) Intravenously (i.v.) administered anti-CD4 antibody was used to distinguish circulating (red pulp localized) versus resident (white pulp localized) CD4+ T cells. (d) Analysis of the GP33+ T cell response 60 days post acute LCMV Armstrong infection. Percentages and numbers of GP33+ T cells in Il10f/f and Il10f/f Foxp3-Cre mice are shown. Data are from one experiment representative of 3 experiments (a, b, c, d) with 3-6 mice per group carried out 45-60 days (a, d), or 0, 8, and 15 days (b, c) following LCMV Armstrong infection (mean and s.e.m).
Supplementary Figure 4 Validation of RNA-seq results.
(a) Expression of Zeb2, Ccr7, Cx3cr1, and Pim1 was determined by qPCR analysis of cDNA isolated from pooled GP33+ and NP396+ CD8+ T cells at 15 days post acute LCMV infection from Il10f/f and Il10f/f Foxp3-Cre mice. Expression of TCF-1 and GzmB in GP33+ in T cells at 15 days post acute LCMV infection from Il10f/f and Il10f/f Foxp3-Cre mice was determined by flow cytometric analysis. (b) Representative plots of the GP33+ T cell response in Il10f/f and Il10f/f Foxp3Cre mice at day 15 post LCMV infection. Data are from one experiment representative of 3 experiments with 3-5 mice per group carried out 15 days following LCMV Armstrong infection (mean and s.e.m).
Supplementary Figure 5 Heat map of differentially expressed genes based on RNA-seq results.
Genes with a p-adjusted value < 0.2 (Benjami-Hochberg) and the corresponding log2 fold-change in mRNA isolated from pooled GP33+ and NP396+ CD8+ T cells at 15 days post acute LCMV infection from Il10f/f and Il10f/f Foxp3-Cre mice.
Supplementary Figure 6 Virus-specific CD8+ T cells from mice lacking CD4+ Treg cell–derived IL-10 display a robust inflammatory and effector gene signature.
mRNA was isolated from pooled GP33+ and NP396+ CD8+ T cells at 15 days post acute LCMV infection from Il10f/f and Il10f/f Foxp3-Cre mice and compared by RNA-seq. (a) Gene set plots showing individual log2 fold-changes of with corresponding standard error based on published effector vs memory gene set. Gene Set Enrichment Analysis (GSEA) was performed using gene sets from the Broad MSigDB collection; select significantly enriched gene sets (FDR < 1e-5) are shown with their running Enrichment Score (ES) (line), where members of the gene set appear in the ranked list of genes (barcode), and the signal to noise ranking metric (bar). A positive ES signifies enrichment in the Il10f/f Foxp3-Cre sample relative to the WT condition of a given gene set; i.e., more highly expressed. (b) GSEA results of CpG (c) and poly:IC stimulated genes (bottom) were visualized. (d) Normalized enrichment scores for Gene Set Enrichment Analysis. Normalized enrichment scores (NES) was calculated for select significantly enriched gene sets (FDR < 1e-5). Gene set name, figure GSEA plots shown in, and NES are shown in table.
Supplementary Figure 7 Transfer of IL-10-competent CD4+ Treg cells during the resolution phase of LCMV infection is sufficient to ‘rescue’ the maturation defect of memory CD8+ T cells in Il10–/– mice.
Analysis of the GP33+ T cell response 60 days p.i. in Foxp3GFP-DTR mice treated with diphtheria toxin at day -1, day 8, or day 15 p.i. or mock injected with PBS. (a) Percentage and numbers of GP33+ T cells are shown. Representative of 3 independent experiments with 3-6 mice per group carried out 45-60 days following LCMV Armstrong infection. (b) Analysis of the GP33+ T cell response 60 days post acute LCMV Armstrong infection in Il10–/– mice and Il10–/– mice that were administered 3x105 Foxp3+ CD4+ T cells isolated from coinfected Foxp3GFP-DTR mice at day 8 p.i. Percentage and numbers of GP33+ T cells are shown. Representative of 2 independent experiments with 3-7 mice per group carried out 60 days following LCMV Armstrong infection. Data are from one experiment representative of 3 experiments (a) or 2 experiments (b) with 3-7 mice per group carried out 45-60 days following LCMV Armstrong infection (mean and s.e.m).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 993 kb)
Rights and permissions
About this article
Cite this article
Laidlaw, B., Cui, W., Amezquita, R. et al. Production of IL-10 by CD4+ regulatory T cells during the resolution of infection promotes the maturation of memory CD8+ T cells. Nat Immunol 16, 871–879 (2015). https://doi.org/10.1038/ni.3224
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3224
This article is cited by
-
B-cell-derived IL-10 promotes allergic sensitization in asthma regulated by Bcl-3
Cellular & Molecular Immunology (2023)
-
The precursors of CD8+ tissue resident memory T cells: from lymphoid organs to infected tissues
Nature Reviews Immunology (2022)
-
Metformin enhances the antitumor activity of oncolytic herpes simplex virus HF10 (canerpaturev) in a pancreatic cell cancer subcutaneous model
Scientific Reports (2022)
-
Bacteroides uniformis CECT 7771 alleviates inflammation within the gut-adipose tissue axis involving TLR5 signaling in obese mice
Scientific Reports (2021)
-
Cord blood levels of interleukin-10 decrease in neonates with increased birth weight: novel implications of the cytokine network in early obesity
European Journal of Pediatrics (2021)