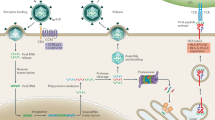
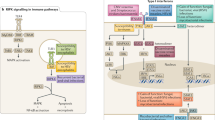
Abstract
The outcome after infection with the human immunodeficiency virus type 1 (HIV-1) is a complex phenotype determined by interactions among the pathogen, the human host and the surrounding environment. An impact of host genetic variation on HIV-1 susceptibility was identified early in the pandemic, with a major role attributed to the genes encoding class I human leukocyte antigens (HLA) and the chemokine receptor CCR5. Studies using genome-wide data sets have underscored the strength of these associations relative to variants located throughout the rest of the genome. However, the extent to which additional polymorphisms influence HIV-1 disease progression, and how much of the variability in outcome can be attributed to host genetics, remain largely unclear. Here we discuss findings concerning the functional impact of associated variants, outline methods for quantifying the host genetic component and examine how available genome-wide data sets may be leveraged to discover gene variants that affect the outcome of HIV-1 infection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Horton, R.E., McLaren, P.J., Fowke, K., Kimani, J. & Ball, T.B. Cohorts for the study of HIV-1-exposed but uninfected individuals: benefits and limitations. J. Infect. Dis. 202 (suppl. 3), S377–S381 (2010).
An, P. & Winkler, C.A. Host genes associated with HIV/AIDS: advances in gene discovery. Trends Genet. 26, 119–131 (2010).
Lane, J. et al. A genome-wide association study of resistance to HIV infection in highly exposed uninfected individuals with hemophilia A. Hum. Mol. Genet. 22, 1903–1910 (2013).
McLaren, P.J. et al. Association study of common genetic variants and HIV-1 acquisition in 6,300 infected cases and 7,200 controls. PLoS Pathog. 9, e1003515 (2013).
Gurdasani, D. et al. A systematic review of definitions of extreme phenotypes of HIV control and progression. AIDS 28, 149–162 (2014).
Kaslow, R.A. et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2, 405–411 (1996).
Carrington, M. et al. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283, 1748–1752 (1999).
Gao, X. et al. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N. Engl. J. Med. 344, 1668–1675 (2001).
Keet, I.P. et al. Consistent associations of HLA class I and II and transporter gene products with progression of human immunodeficiency virus type 1 infection in homosexual men. J. Infect. Dis. 180, 299–309 (1999).
Martin, M.P. & Carrington, M. Immunogenetics of HIV disease. Immunol. Rev. 254, 245–264 (2013).
Fellay, J. et al. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 5, e1000791 (2009).
Pereyra, F. et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330, 1551–1557 (2010).
Carrington, M., Bashirova, A.A. & McLaren, P.J. On stand by: host genetics of HIV control. AIDS 27, 2831–2839 (2013).
Pyo, C.W. et al. Recombinant structures expand and contract inter and intragenic diversification at the KIR locus. BMC Genomics 14, 89 (2013).
Fellay, J. et al. A whole-genome association study of major determinants for host control of HIV-1. Science 317, 944–947 (2007). This is the first genome-wide association study on an infectious disease trait. This study demonstrates that variation in the MHC region is the primary host genetic influence on HIV outcome in an unbiased, genome-wide screen.
Migueles, S.A. et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long-term nonprogressors. Proc. Natl. Acad. Sci. USA 97, 2709–2714 (2000).
Jia, X. et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS ONE 8, e64683 (2013).
Dunstan, S.J. et al. Variation at HLA-DRB1 is associated with resistance to enteric fever. Nat. Genet. 46, 1333–1336 (2014).
Patsopoulos, N.A. et al. Fine-mapping the genetic association of the major histocompatibility complex in multiple sclerosis: HLA and non-HLA effects. PLoS Genet. 9, e1003926 (2013).
Okada, Y. et al. Fine mapping major histocompatibility complex associations in psoriasis and its clinical subtypes. Am. J. Hum. Genet. 95, 162–172 (2014).
Raychaudhuri, S. et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet. 44, 291–296 (2012).
Stranger, B.E. et al. Population genomics of human gene expression. Nat. Genet. 39, 1217–1224 (2007).
Thomas, R. et al. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat. Genet. 41, 1290–1294 (2009).
Kaufman, J. & Salomonsen, J. The “minimal essential MHC” revisited: both peptide-binding and cell surface expression level of MHC molecules are polymorphisms selected by pathogens in chickens. Hereditas 127, 67–73 (1997).
Koch, M. et al. Structures of an MHC class I molecule from B21 chickens illustrate promiscuous peptide binding. Immunity 27, 885–899 (2007).
Corrah, T.W. et al. Reappraisal of the relationship between the HIV-1-protective single-nucleotide polymorphism 35 kilobases upstream of the HLA-C gene and surface HLA-C expression. J. Virol. 85, 3367–3374 (2011).
Pulit, S.L., Voight, B.F. & de Bakker, P.I. Multiethnic genetic association studies improve power for locus discovery. PLoS ONE 5, e12600 (2010).
Apps, R. et al. Influence of HLA-C expression level on HIV control. Science 340, 87–91 (2013). The authors determine the cell surface expression dynamics of all common classical HLA-C allotypes . Accounting for linkage disequilibrium with known allelic effects at HLA-A and HLA-B, they demonstrate an independent impact of HLA-C expression level on HIV control.
Kulkarni, S. et al. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature 472, 495–498 (2011).
Kulkarni, S. et al. Genetic interplay between HLA-C and MIR148A in HIV control and Crohn disease. Proc. Natl. Acad. Sci. USA 110, 20705–20710 (2013).
Blais, M.E. et al. High frequency of HIV mutations associated with HLA-C suggests enhanced HLA-C-restricted CTL selective pressure associated with an AIDS-protective polymorphism. J. Immunol. 188, 4663–4670 (2012).
Kiepiela, P. et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432, 769–775 (2004).
Kiepiela, P. et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13, 46–53 (2007).
Petersdorf, E.W. et al. HLA-C expression levels define permissible mismatches in hematopoietic cell transplantation. Blood 124, 3996–4003 (2014).
Thomas, R. et al. A novel variant marking HLA-DP expression levels predicts recovery from hepatitis B virus infection. J. Virol. 86, 6979–6985 (2012).
Duggal, P. et al. Genome-wide association study of spontaneous resolution of hepatitis C virus infection: data from multiple cohorts. Ann. Intern. Med. 158, 235–245 (2013).
Wissemann, W.T. et al. Association of Parkinson disease with structural and regulatory variants in the HLA region. Am. J. Hum. Genet. 93, 984–993 (2013).
Reits, E.A. et al. Radiation modulates the peptide repertoire, enhances MHC class I expression, and induces successful antitumor immunotherapy. J. Exp. Med. 203, 1259–1271 (2006).
Faroudi, M. et al. Lytic versus stimulatory synapse in cytotoxic T lymphocyte/target cell interaction: manifestation of a dual activation threshold. Proc. Natl. Acad. Sci. USA 100, 14145–14150 (2003).
Stahl, E.A. et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat. Genet. 44, 483–489 (2012).
Yang, J. et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–569 (2010). Missing heritabililty is the proportion of trait variability known to be attributable to genetic effects that has not been explained by GWAS. Using human height as a model, the authors describe a method for quantifying the heritability explained by all common SNPs and show that this accounts for a large fraction of the missing heritability.
Gusev, A. et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am. J. Hum. Genet. 95, 535–552 (2014).
Carlson, J.M. et al. HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science 345, 1254031 doi:10.1126/science.1254031 (2014).
Fraser, C. et al. Virulence and pathogenesis of HIV-1 infection: an evolutionary perspective. Science 343, 1243727 doi:10.1126/science.1243727 (2014).
Fraser, C., Hollingsworth, T.D., Chapman, R., de Wolf, F. & Hanage, W.P. Variation in HIV-1 set-point viral load: epidemiological analysis and an evolutionary hypothesis. Proc. Natl. Acad. Sci. USA 104, 17441–17446 (2007).
van Dorp, C.H., van Boven, M. & de Boer, R.J. Immuno-epidemiological modeling of HIV-1 predicts high heritability of the set-point virus load, while selection for CTL escape dominates virulence evolution. PLoS Comput. Biol. 10, e1003899 (2014).
Bartha, I. et al. A genome-to-genome analysis of associations between human genetic variation, HIV-1 sequence diversity, and viral control. Elife 2, e01123 (2013). In a combined analysis of host and pathogen genetics, the authors use HIV sequence diversity as phenotype in multiple GWAS. The authors demonstrated that using sites of viral escape as an intermediate phenotype was more powerful than using clinical markers of disease progression to detect HLA associations.
Mackelprang, R.D. et al. Host genetic and viral determinants of HIV-1 RNA set-point among HIV-1 seroconverters from sub-Saharan Africa. J. Virol. 89, 2104–2111 (2015).
Frater, A.J. et al. Effective T-cell responses select human immunodeficiency virus mutants and slow disease progression. J. Virol. 81, 6742–6751 (2007).
Leslie, A.J. et al. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10, 282–289 (2004).
Martinez-Picado, J. et al. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 80, 3617–3623 (2006).
Schneidewind, A. et al. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J. Virol. 81, 12382–12393 (2007).
Kawashima, Y. et al. Adaptation of HIV-1 to human leukocyte antigen class I. Nature 458, 641–645 (2009). In a study of HIV diversity in multiple geographic regions, the authors show a correlation between frequency of viral escape mutations and the restricting HLA alleles. This adaptation resulted in the loss of protective effect of HLA-B*51 in a Japanese sample over time.
Cotton, L.A. et al. Genotypic and functional impact of HIV-1 adaptation to its host population during the North American epidemic. PLoS Genet. 10, e1004295 (2014).
Payne, R. et al. Impact of HLA-driven HIV adaptation on virulence in populations of high HIV seroprevalence. Proc. Natl. Acad. Sci. USA 111, E5393–E5400 (2014).
Herbeck, J.T. et al. Is the virulence of HIV changing? A meta-analysis of trends in prognostic markers of HIV disease progression and transmission. AIDS 26, 193–205 (2012).
Regoes, R.R. et al. Disentangling human tolerance and resistance against HIV. PLoS Biol. 12, e1001951 (2014).
Abecasis, G.R. et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Jägger, S. et al. Global landscape of HIV–human protein complexes. Nature 481, 365–370 (2012).
Zhu, J. et al. Comprehensive identification of host modulators of HIV-1 replication using multiple orthologous RNAi reagents. Cell Reports 9, 752–766 (2014).
Akashi, H., Osada, N. & Ohta, T. Weak selection and protein evolution. Genetics 192, 15–31 (2012).
Farzadegan, H. et al. Sex differences in HIV-1 viral load and progression to AIDS. Lancet 352, 1510–1514 (1998).
Sterling, T.R. et al. Sex differences in longitudinal human immunodeficiency virus type 1 RNA levels among seroconverters. J. Infect. Dis. 180, 666–672 (1999).
Konttinen, Y.T., Hanninen, A. & Fuellen, G. Plasmacytoid dendritic cells, Janus-faced sentinels: progesterone, guilty or innocent? Immunotherapy 1, 929–931 (2009).
Kiezun, A. et al. Exome sequencing and the genetic basis of complex traits. Nat. Genet. 44, 623–630 (2012).
MacArthur, D.G. et al. Guidelines for investigating causality of sequence variants in human disease. Nature 508, 469–476 (2014).
Chang, J. et al. Polymorphisms in interferon regulatory factor 7 reduce interferon-alpha responses of plasmacytoid dendritic cells to HIV-1. AIDS 25, 715–717 (2011).
Lodi, S. et al. Immunovirologic control 24 months after interruption of antiretroviral therapy initiated close to HIV seroconversion. Arch. Intern. Med. 172, 1252–1255 (2012).
Sáez-Cirión, A. et al. Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS Pathog. 9, e1003211 (2013).
Ge, D. et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461, 399–401 (2009).
Prentice, H.A. et al. HLA class I, KIR, and genome-wide SNP diversity in the RV144 Thai phase 3 HIV vaccine clinical trial. Immunogenetics 66, 299–310 (2014).
Deeks, S.G. & Walker, B.D. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity 27, 406–416 (2007).
Bartha, I., McLaren, P.J., Ciuffi, A., Fellay, J. & Telenti, A. GuavaH: a compendium of host genomic data in HIV biology and disease. Retrovirology 11, 6 (2014).
Acknowledgements
We thank A. Bashirova and I. Bartha for helpful suggestions. This project was funded in whole or in part with federal funds from the Frederick National Laboratory for Cancer Research, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government. This research was supported in part by the Intramural Research Program of the National Institutes of Health, Frederick National Laboratory, Center for Cancer Research.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
McLaren, P., Carrington, M. The impact of host genetic variation on infection with HIV-1. Nat Immunol 16, 577–583 (2015). https://doi.org/10.1038/ni.3147
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3147
This article is cited by
-
Assessment of NKG2C copy number variation in HIV-1 infection susceptibility, and considerations about the potential role of lacking receptors and virus infection
Journal of Human Genetics (2022)
-
Genome-wide study of a Neolithic Wartberg grave community reveals distinct HLA variation and hunter-gatherer ancestry
Communications Biology (2021)
-
Distinct viral reservoirs in individuals with spontaneous control of HIV-1
Nature (2020)
-
Human Genetic Variation and HIV/AIDS in Papua New Guinea: Time to Connect the Dots
Current HIV/AIDS Reports (2018)
-
The tumour suppressor APC promotes HIV-1 assembly via interaction with Gag precursor protein
Nature Communications (2017)