Abstract
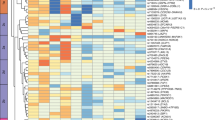
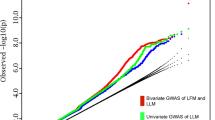
Genome-wide association studies have identified 32 loci influencing body mass index, but this measure does not distinguish lean from fat mass. To identify adiposity loci, we meta-analyzed associations between ∼2.5 million SNPs and body fat percentage from 36,626 individuals and followed up the 14 most significant (P < 10−6) independent loci in 39,576 individuals. We confirmed a previously established adiposity locus in FTO (P = 3 × 10−26) and identified two new loci associated with body fat percentage, one near IRS1 (P = 4 × 10−11) and one near SPRY2 (P = 3 × 10−8). Both loci contain genes with potential links to adipocyte physiology. Notably, the body-fat–decreasing allele near IRS1 is associated with decreased IRS1 expression and with an impaired metabolic profile, including an increased visceral to subcutaneous fat ratio, insulin resistance, dyslipidemia, risk of diabetes and coronary artery disease and decreased adiponectin levels. Our findings provide new insights into adiposity and insulin resistance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Anonymous. Obesity: preventing and managing the global epidemic. Report of a WHO consultation. World Health Organ. Tech. Rep. Ser. 894, 1–253 (2000).
Maes, H.H., Neale, M.C. & Eaves, L.J. Genetic and environmental factors in relative body weight and human adiposity. Behav. Genet. 27, 325–351 (1997).
Frayling, T.M. et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316, 889–894 (2007).
Loos, R.J. et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 40, 768–775 (2008).
Scuteri, A. et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 3, e115 (2007).
Thorleifsson, G. et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet. 41, 18–24 (2009).
Willer, C.J. et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 41, 25–34 (2009).
Speliotes, E.K. et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 42, 937–948 (2010).
Teslovich, T.M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010).
Rung, J. et al. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat. Genet. 41, 1110–1115 (2009).
Samani, N.J. et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 357, 443–453 (2007).
Matsuda, M. & DeFronzo, R.A. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care 22, 1462–1470 (1999).
Gutt, M. et al. Validation of the insulin sensitivity index (ISI(0,120)): comparison with other measures. Diabetes Res. Clin. Pract. 47, 177–184 (2000).
Badman, M.K. & Flier, J.S. The adipocyte as an active participant in energy balance and metabolism. Gastroenterology 132, 2103–2115 (2007).
Kim, J.Y. et al. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J. Clin. Invest. 117, 2621–2637 (2007).
Virtue, S. & Vidal-Puig, A. It's not how fat you are, it's what you do with it that counts. PLoS Biol. 6, e237 (2008).
Emilsson, V. et al. Genetics of gene expression and its effect on disease. Nature 452, 423–428 (2008).
Zhong, H., Yang, X., Kaplan, L.M., Molony, C. & Schadt, E.E. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am. J. Hum. Genet. 86, 581–591 (2010).
Myers, A.J. et al. A survey of genetic human cortical gene expression. Nat. Genet. 39, 1494–1499 (2007).
Cabrita, M.A. & Christofori, G. Sprouty proteins, masterminds of receptor tyrosine kinase signaling. Angiogenesis 11, 53–62 (2008).
Yigzaw, Y., Cartin, L., Pierre, S., Scholich, K. & Patel, T.B. The C terminus of sprouty is important for modulation of cellular migration and proliferation. J. Biol. Chem. 276, 22742–22747 (2001).
Lee, C.C. et al. Overexpression of sprouty 2 inhibits HGF/SF-mediated cell growth, invasion, migration, and cytokinesis. Oncogene 23, 5193–5202 (2004).
Zhang, C. et al. Regulation of vascular smooth muscle cell proliferation and migration by human sprouty 2. Arterioscler. Thromb. Vasc. Biol. 25, 533–538 (2005).
Urs, S. et al. Sprouty1 is a critical regulatory switch of mesenchymal stem cell lineage allocation. FASEB J. 24, 3264–3273 (2010).
Heard-Costa, N.L. et al. NRXN3 is a novel locus for waist circumference: a genome-wide association study from the CHARGE Consortium. PLoS Genet. 5, e1000539 (2009).
Lindgren, C.M. et al. Genome-wide association scan meta-analysis identifies three loci influencing adiposity and fat distribution. PLoS Genet. 5, e1000508 (2009).
Meyre, D. et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat. Genet. 41, 157–159 (2009).
Scherag, A. et al. Two new loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and German study groups. PLoS Genet. 6, e1000916 (2010).
Heid, I.M. et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 42, 949–960 (2010).
Grarup, N., Sparso, T. & Hansen, T. Physiologic characterization of type 2 diabetes-related loci. Curr. Diab. Rep. 10, 485–497 (2010).
Stumvoll, M. & Jacob, S. Multiple sites of insulin resistance: muscle, liver and adipose tissue. Exp. Clin. Endocrinol. Diabetes 107, 107–110 (1999).
Arner, P. Insulin resistance in type 2 diabetes: role of fatty acids. Diabetes Metab. Res. Rev. 18 (Suppl 2) S5–S9 (2002).
Wajchenberg, B.L. Subcutaneous and visceral adipose tissue: their relation to the metabolic syndrome. Endocr. Rev. 21, 697–738 (2000).
Araki, E. et al. Alternative pathway of insulin signalling in mice with targeted disruption of the IRS-1 gene. Nature 372, 186–190 (1994).
Tamemoto, H. et al. Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Nature 372, 182–186 (1994).
Zhou, L. et al. Insulin receptor substrate-2 (IRS-2) can mediate the action of insulin to stimulate translocation of GLUT4 to the cell surface in rat adipose cells. J. Biol. Chem. 272, 29829–29833 (1997).
Kaburagi, Y. et al. Role of insulin receptor substrate-1 and pp60 in the regulation of insulin-induced glucose transport and GLUT4 translocation in primary adipocytes. J. Biol. Chem. 272, 25839–25844 (1997).
Smith-Hall, J. et al. The 60 kDa insulin receptor substrate functions like an IRS protein (pp60IRS3) in adipose cells. Biochemistry 36, 8304–8310 (1997).
Laustsen, P.G. et al. Lipoatrophic diabetes in Irs1(−/−)/Irs3(−/−) double knockout mice. Genes Dev. 16, 3213–3222 (2002).
Miki, H. et al. Essential role of insulin receptor substrate 1 (IRS-1) and IRS-2 in adipocyte differentiation. Mol. Cell. Biol. 21, 2521–2532 (2001).
Tseng, Y.H., Kriauciunas, K.M., Kokkotou, E. & Kahn, C.R. Differential roles of insulin receptor substrates in brown adipocyte differentiation. Mol. Cell. Biol. 24, 1918–1929 (2004).
Reich, D., Thangaraj, K., Patterson, N., Price, A.L. & Singh, L. Reconstructing Indian population history. Nature 461, 489–494 (2009).
Barnett, A.H. et al. Type 2 diabetes and cardiovascular risk in the UK south Asian community. Diabetologia 49, 2234–2246 (2006).
Pruim, R.J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010).
Li, Y. & Mach Abecasis, G.R. 1.0: rapid haplotype reconstruction and missing genotype inference. Am. J. Hum. Genet. S79, 2290 (2006).
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913 (2007).
Servin, B. & Stephens, M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 3, e114 (2007).
Chambers, J.C. et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat. Genet. 40, 716–718 (2008).
Abecasis, G.R., Cherny, S.S., Cookson, W.O. & Cardon, L.R. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30, 97–101 (2002).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Wang, Z.V., Deng, Y., Wang, Q.A., Sun, K. & Scherer, P.E. Identification and characterization of a promoter cassette conferring adipocyte-specific gene expression. Endocrinology 151, 2933–2939 (2010).
Acknowledgements
A full list of Acknowledgments appears in the Supplementary Note. Funding was provided by Academy of Finland (10404, 124243, 129680, 129494, 141005 and 213506); Agency for Health Care Policy Research (HS06516); Althingi (the Icelandic Parliament); American Heart Association (10SDG269004); AstraZeneca; Baltimore Geriatric Research Education and Clinical Centers; Biocentrum Helsinki Foundation; Biotechnology and Biological Sciences Research Council (G20234); British Heart Foundation (PG/07/133/24260, RG/08/008, SP/04/002, SP/07/007/23671); CamStrad; Cancer Research UK; Cedars-Sinai Board of Governors' Chair in Medical Genetics; Centre for Medical Systems Biology (The Netherlands); Centre Hospitalier Universitaire Vaudois (Lausanne); Croatian Ministry of Science, Education and Sport (196-1962766-2747, 216-1080315-0302 and 309-0061194-2023); Department of Health (UK); Department of Veterans Affairs (USA); Emil and Vera Cornell Foundation; Erasmus Medical Center (Rotterdam); Erasmus University (Rotterdam); European Commission (DG XII, FP7/2007-2013, FP7-KBBE-2010-4-266408, HEALTH-F2-2007-201681, HEALTH-F2-2008-201865-GEFOS, HEALTH-F4-2007-201413, HEALTH-F4-2007-201550, LSHG-CT-2006-018947, LSHG-CT-2006-01947, LSHM-CT-2003-503041, LSHM-CT-2004-512013, QLG1-CT-2001-01252 and QLG2-CT-2002-01254); Finnish Diabetes Foundation; Finnish Diabetes Research Foundation; Finnish Foundation for Cardiovascular Research; Finnish Heart Foundation; Finnish Medical Society; Folkhälsan Research Foundation; Food Standards Agency (UK); Foundation for Life and Health in Finland; German Bündesministerium für Forschung und Technology (01AK803A-H and 01IG07015G); German Federal Ministry of Education and Research; German National Genome Research Network (NGFN-2 and NGFNPlus: 01GS0823); Giorgi-Cavaglieri Foundation; GlaxoSmithKline; Göteborg Medical Society; Gyllenberg Foundation; Health and Safety Executive (UK); Health Care Centers in Vasa, Närpes and Korsholm; Hjartavernd (the Icelandic Heart Association); John D. and Catherine T. MacArthur Foundation; Knut and Alice Wallenberg Foundation; Leenaards Foundation; Ludwig-Maximilians Universität München; Lundberg Foundation; Medical Research Council (UK); Men's Associates of Hebrew SeniorLife; Ministerio de Ciencia e Innovación (Spain) (SAF-2009 and SAF-2008-02073); Ministry for Health, Welfare and Sports (The Netherlands); Ministry of Education (Finland); Ministry of Education, Culture and Science (The Netherlands); Municipal Health Care Center and Hospital in Jakobstad; Municipality of Rotterdam; Närpes Health Care Foundation; National Institute for Health Research (UK); US National Institutes of Health (USA) (AG13196, DK063491, K23-DK080145, M01-RR00425, N01-AG12100, N01-AG62101, N01-AG62103, N01-AG62106, N01-HC15103, N01-HC25195, N01-HC35129, N01-HC45133, N01-HC55222, N01-HC75150, N01-HC85079 through N01-HC85086, P30-DK072488, R01-AG031890-01, R01-AG18728, R01-AG032098-01A1, R01-AR/AG41398, R01-AR046838, R01-DK06833603, R01-DK075787, R01-DK07568102, R01-HL036310-20A2, R01-HL087652, R01-HL08770003, R01-HL088119, U01-HL080295, U01-HL72515 and U01-HL84756); Netherlands Genomics Initiative/Netherlands Consortium for Healthy Aging (050-060-810); Netherlands Organisation for Scientific Research (175.010.2005.011 and 911-03-012); Netherlands Organization for the Health Research and Development; Nordic Center of Excellence in Disease Genetics; Novo Nordisk Foundation; Ollqvist Foundation; Paavo Nurmi Foundation; Perklén Foundation; Petrus and Augusta Hedlunds Foundation; Research Institute for Diseases in the Elderly (014-93-015; RIDE2); Robert Dawson Evans Endowment; Royal Society (UK); Sahlgrenska Center for Cardiovascular and Metabolic Research (A305:188); Sahlgrenska University Hospital Foundation (ALF/LUA); Science Funding programme (UK); Scottish Executive Health Department; Sigrid Juselius Foundation; State of Bavaria; Stroke Association (UK); Swedish Cultural Foundation in Finland; Swedish Foundation for Strategic Research; Swedish Research Council (K2010-54X-09894-19-3, K2010-52X-20229-05-3 and 2006-3832); Swedish Strategic Foundation; Swiss Institute of Bioinformatics; Swiss National Science Foundation (3100AO-116323/1, 310000-112552 and 33CSCO-122661); TEKES (1510/31/06); Torsten and Ragnar Söderberg's Foundation; United Kingdom NIHR Cambridge Biomedical Research Centre; University of Lausanne; University of Maryland General Clinical Research Center (M01 RR 16500); Uppsala University; Västra Götaland Foundation; and Wellcome Trust (077016/Z/05/Z, 084723/Z/08/Z and 091746/Z/10/Z).
Author information
Authors and Affiliations
Contributions
A full list of author contributions appears in the Supplementary Note.
Corresponding author
Ethics declarations
Competing interests
I.B. and spouse own stock in Incyte Ltd and GlaxoSmithKline. K.S., G.T., U.T. and U.S. are employed by deCODE Genetics. V.M. is a full-time employee of GlaxoSmithKline. G.W. and P.V. received funding from GlaxoSmithKline to build the CoLaus Study.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–9, Supplementary Figures 1–7 and Supplementary Note. (PDF 1136 kb)
Rights and permissions
About this article
Cite this article
Kilpeläinen, T., Zillikens, M., Stančákova, A. et al. Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile. Nat Genet 43, 753–760 (2011). https://doi.org/10.1038/ng.866
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.866
This article is cited by
-
Sex differences in energy metabolism: natural selection, mechanisms and consequences
Nature Reviews Nephrology (2024)
-
Transcriptome-wide association study-derived genes as potential visceral adipose tissue-specific targets for type 2 diabetes
Diabetologia (2023)
-
Age effect on the shared etiology of glycemic traits and serum lipids: evidence from a Chinese twin study
Journal of Endocrinological Investigation (2023)
-
Genetics and Epigenetics in Obesity: What Do We Know so Far?
Current Obesity Reports (2023)
-
A non-coding variant linked to metabolic obesity with normal weight affects actin remodelling in subcutaneous adipocytes
Nature Metabolism (2023)