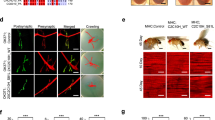
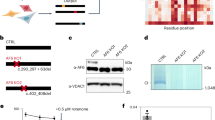
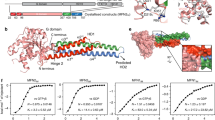
Abstract
Although mutations in CYTB (cytochrome b) or BCS1L have been reported in isolated defects of mitochondrial respiratory chain complex III (cIII), most cIII-defective individuals remain genetically undefined. We identified a homozygous nonsense mutation in the gene encoding tetratricopeptide 19 (TTC19) in individuals from two families affected by progressive encephalopathy associated with profound cIII deficiency and accumulation of cIII-specific assembly intermediates. We later found a second homozygous nonsense mutation in a fourth affected individual. We demonstrated that TTC19 is embedded in the inner mitochondrial membrane as part of two high–molecular‐weight complexes, one of which coincides with cIII. We then showed a physical interaction between TTC19 and cIII by coimmunoprecipitation. We also investigated a Drosophila melanogaster knockout model for TTC19 that showed low fertility, adult-onset locomotor impairment and bang sensitivity, associated with cIII deficiency. TTC19 is a putative cIII assembly factor whose disruption is associated with severe neurological abnormalities in humans and flies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Iwata, S. et al. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science 281, 64–71 (1998).
Schägger, H., Link, T.A., Engel, W.D. & von Jagow, G. Isolation of the eleven protein subunits of the bc1 complex from beef heart. Methods Enzymol. 126, 224–237 (1986).
Wittig, I., Braun, H.P. & Schagger, H. Blue native PAGE. Nat. Protoc. 1, 418–428 (2006).
Fernandez-Vizarra, E. et al. Impaired complex III assembly associated with BCS1L gene mutations in isolated mitochondrial encephalopathy. Hum. Mol. Genet. 16, 1241–1252 (2007).
Wimplinger, I. et al. Mutations of the mitochondrial holocytochrome c-type synthase in X-linked dominant microphthalmia with linear skin defects syndrome. Am. J. Hum. Genet. 79, 878–889 (2006).
Pagliarini, D.J. et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 (2008).
Sagona, A.P. et al. PtdIns(3)P controls cytokinesis through KIF13A mediated recruitment of FYVE-CENT to the midbody. Nat. Cell Biol. 12, 362–371 (2010).
Brandt, U., Yu, L., Yu, C.A. & Trumpower, B.L. The mitochondrial targeting presequence of the Rieske iron-sulfur protein is processed in a single step after insertion into the cytochrome bc1 complex in mammals and retained as a subunit in the complex. J. Biol. Chem. 268, 8387–8390 (1993).
Cruciat, C.M., Hell, K., Folsch, H., Neupert, W. & Stuart, R.A. Bcs1p, an AAA-family member, is a chaperone for the assembly of the cytochrome bc(1) complex. EMBO J. 18, 5226–5233 (1999).
Zara, V., Palmisano, I., Conte, L. & Trumpower, B.L. Further insights into the assembly of the yeast cytochrome bc1 complex based on analysis of single and double deletion mutants lacking supernumerary subunits and cytochrome b. Eur. J. Biochem. 271, 1209–1218 (2004).
Lightowlers, R.N. & Chrzanowska-Lightowlers, Z.M. PPR (pentatricopeptide repeat) proteins in mammals: important aids to mitochondrial gene expression. Biochem. J. 416, e5–e6 (2008).
Small, I.D. & Peeters, N. The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 46–47 (2000).
Giansanti, M.G. et al. Genetic dissection of meiotic cytokinesis in Drosophila males. Mol. Biol. Cell 15, 2509–2522 (2004).
Glasscock, E. & Tanouye, M.A. Drosophila couch potato mutants exhibit complex neurological abnormalities including epilepsy phenotypes. Genetics 169, 2137–2149 (2005).
Zordan, M.A. et al. Post-transcriptional silencing and functional characterization of the Drosophila melanogaster homolog of human Surf1. Genetics 172, 229–241 (2006).
Borst, A., Haag, J. & Rieff, D.F. Fly motion vision. Annu. Rev. Neurosci. 33, 49–70 (2010).
Minai, L. et al. Mitochondrial respiratory chain complex assembly and function during human fetal development. Mol. Genet. Metab. 94, 120–126 (2008).
Blatch, G.L. & Lassle, M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21, 932–939 (1999).
Kimmins, S. & MacRae, T.H. Maturation of steroid receptors: an example of functional cooperation among molecular chaperones and their associated proteins. Cell Stress Chaperones 5, 76–86 (2000).
Kotarsky, H. et al. BCS1L is expressed in critical regions for neural development during ontogenesis in mice. Gene Expr. Patterns 7, 266–273 (2007).
Papa, S. Mitochondrial oxidative phosphorylation changes in the life span. Molecular aspects and physiopathological implications. Biochim. Biophys. Acta 1276, 87–105 (1996).
Bugiani, M. et al. Clinical and molecular findings in children with complex I deficiency. Biochim. Biophys. Acta 1659, 136–147 (2004).
Wu, M. et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am. J. Physiol. Cell Physiol. 292, C125–C136 (2007).
Tiranti, V. et al. Characterization of SURF-1 expression and Surf-1p function in normal and disease conditions. Hum. Mol. Genet. 8, 2533–2540 (1999).
Zhang, J.C. et al. Down-regulation of CXCR4 expression by SDF-KDEL in CD34(+) hematopoietic stem cells: an antihuman immunodeficiency virus strategy. J. Virol. Methods 16, 30–37 (2009).
Ghezzi, D. et al. Paroxysmal non-kinesigenic dyskinesia is caused by mutations of the MR-1 mitochondrial targeting sequence. Hum. Mol. Genet. 18, 1058–1064 (2009).
Heckmatt, J.Z. & Dubowitz, V. Needle biopsy of skeletal muscle. Muscle Nerve 7, 594 (1984).
Sciacco, M. & Bonilla, E. Cytochemistry and immunocytochemistry of mitochondria in tissue sections. Methods Enzymol. 264, 509–521 (1996).
Tiranti, V. et al. Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 63, 1609–1621 (1998).
Nijtmans, L.G., Henderson, N.S. & Holt, I.J. Blue native electrophoresis to study mitochondrial and other protein complexes. Methods 26, 327–334 (2002).
Schägger, H. & von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379 (1987).
Fernández-Silva, P., Acin-Perez, R., Fernandez-Vizarra, E., Perez-Martos, A. & Enriquez, J.A. In vivo and in organello analyses of mitochondrial translation. Methods Cell Biol. 80, 571–588 (2007).
Chomyn, A. In vivo labeling and analysis of human mitochondrial translation products. Methods Enzymol. 264, 197–211 (1996).
Parks, A.L. et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36, 288–292 (2004).
Dietzl, G. et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156 (2007).
Acknowledgements
We are grateful to S. Zanotti for technical support in preparing myoblast cultures and R. Saccon and D. Zanini for helping with the locomotor assays and bang tests. We thank M. Gatti, 'La Sapienza' University, Rome, Italy, for helping with analysis of spermatid development in D. melanogaster and A. Megighian for ERGs studies. The EuroBioBank and Telethon Network of Genetic Biobanks (GTB07001F grant to M. Mora) are also gratefully acknowledged for providing biological samples. This work was supported by the Pierfranco and Luisa Mariani Foundation Italy, the Fondazione Telethon-Italy grant numbers GGP07019 and GPP10005, and grant RF-INN-2007-634163 of the Italian Ministry of Health to M. Zeviani and by grants of the Cariparo Foundation; the Ministry of Education and Research and Progetto Strategico Università di Padova 'Models of Mitochondrial Disease' to R.C.
Author information
Authors and Affiliations
Contributions
D.G. and P.A. found TTC19 and characterized the mutations in human cells. C.L. performed the histological analysis of muscle biopsies. M. Zordan, C.D.R., C.B. and R.C. carried out the experiments in flies. C.M., G.U. and C.S. identified the subjects and carried out the clinical workout. P.D'A. performed linkage analysis. D.D. carried out the mutational screening on subjects 3 and 4 and the controls. M. Zeviani conceived the experimental planning and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1–12 and Supplementary Tables 1–5 (PDF 6898 kb)
Rights and permissions
About this article
Cite this article
Ghezzi, D., Arzuffi, P., Zordan, M. et al. Mutations in TTC19 cause mitochondrial complex III deficiency and neurological impairment in humans and flies. Nat Genet 43, 259–263 (2011). https://doi.org/10.1038/ng.761
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.761
This article is cited by
-
Mitochondria regulate intracellular coenzyme Q transport and ferroptotic resistance via STARD7
Nature Cell Biology (2023)
-
Calpain-mediated protein targets in cardiac mitochondria following ischemia–reperfusion
Scientific Reports (2022)
-
The assembly, regulation and function of the mitochondrial respiratory chain
Nature Reviews Molecular Cell Biology (2022)
-
Clinical, imaging, biochemical and molecular features in Leigh syndrome: a study from the Italian network of mitochondrial diseases
Orphanet Journal of Rare Diseases (2021)
-
Exploiting pyocyanin to treat mitochondrial disease due to respiratory complex III dysfunction
Nature Communications (2021)