Abstract
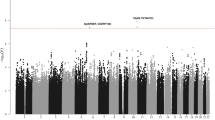
To identify susceptibility loci for non-Hodgkin lymphoma subtypes, we conducted a three-stage genome-wide association study. We identified two variants associated with follicular lymphoma at 6p21.32 (rs10484561, combined P = 1.12 × 10−29 and rs7755224, combined P = 2.00 × 10−19; r2 = 1.0), supporting the idea that major histocompatibility complex genetic variation influences follicular lymphoma susceptibility. We also found confirmatory evidence of a previously reported association between chronic lymphocytic leukemia/small lymphocytic lymphoma and rs735665 (combined P = 4.24 × 10−9).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Skibola, C.F., Curry, J.D. & Nieters, A. Haematologica 92, 960–969 (2007).
Skibola, C.F. et al. Nat. Genet. 41, 873–875 (2009).
Skibola, C.F. et al. PLoS One 3, e2816 (2008).
Smedby, K.E. et al. J. Natl. Cancer Inst. 97, 199–209 (2005).
Cerhan, J.R. et al. Blood 110, 4455–4463 (2007).
Di Bernardo, M.C. et al. Nat. Genet. 40, 1204–1210 (2008).
de Bakker, P.I. et al. Nat. Genet. 38, 1166–1172 (2006).
Klitz, W. et al. Tissue Antigens 62, 296–307 (2003).
Al-Tonbary, Y. et al. Hematology 9, 139–145 (2004).
Nathalang, O. et al. Eur. J. Immunogenet. 26, 389–392 (1999).
Choi, H.B. et al. Int. J. Hematol. 87, 203–209 (2008).
Skibola, C.F. et al. Am. J. Epidemiol. 171, 267–276 (2010).
Abdou, A.M. et al. Leukemia 24, 1055–1058 (2010).
Acknowledgements
This work was supported by grants CA122663 and CA104682 from the National Cancer Institute (NCI), US National Institutes of Health (NIH) (C.F.S.); grants CA45614 and CA89745 from the NCI, NIH (E.A.H.); the American Cancer Society (IRG-07-06401) and a charitable donation by S. Chase (K.B.). E.H. is a faculty fellow of the Edmond J. Safra Bioinformatics program at Tel-Aviv University. SCALE study: We thank X.Y. Chen, H.B. Toh, K.K. Heng and W.Y. Meah from Genome Institute of Singapore for their support in genotyping analyses. We are also grateful to L. Klareskog, Center for Molecular Medicine, and L. Alfredsson, Institute of Environmental Medicine at the Karolinska Institute, Stockholm, Sweden, for sharing DNA from their EIRA study control population and to E. Rehnberg for help with imputing genotypes in the SCALE study. The following agencies contributed funding to SCALE that facilitated the present project: Agency for Science and Technology and Research of Singapore (A*STAR), NCI, the Swedish Cancer Society, the Swedish Research Council and the Danish Medical Research Council. GEC-Mayo Study: This study is supported by grants R01 CA91253 and R01 CA118444 from the NCI. NCI-SEER study: We thank P. Hui of the Information Management Services, Inc. for programming support, and we acknowledge the contributions of the staff and scientists at the SEER centers of Iowa, Los Angeles, Detroit and Seattle for conducting the study's field effort. We especially acknowledge the contributions of study site principal investigator L. Bernstein (Los Angeles). The NCI-SEER study was supported by the Intramural Research Program of the NIH (NCI) and by US Public Health Service (PHS) contracts N01-PC-65064, N01-PC-67008, N01-PC-67009, N01-PC-67010 and N02-PC-71105. Yale study: This study is supported by grant CA62006 from the NCI and the Intramural Research Program of the NIH (NCI). NSW study: The NSW study was supported by a National Health and Medical Research Council of Australia project grant, Cancer Council NSW, a University of Sydney Medical Foundation Program Grant and the Intramural Research Program of the NIH (NCI). BC study: Funding for the British Columbia study was from the Canadian Cancer Society and the Canadian Institutes of Health Research. A.B.-W. is a Senior Scholar of the Michael Smith Foundation for Health Research. EpiLymph study: Funding was available from the German José Carreras Leukemia Foundation (DJCLS_R04/08 and R07/26f) (A.N.), the EC 5th Framework Program Quality of Life grant No. QLK4-CT-2000-00422 (P. Boffetta, P. Brennan), the Federal Office for Radiation Protection grants No. StSch4261 and StSch4420 (Germany), the Spanish Ministry of Health grants Centro de Investigación Biomédica en Red de Epidemiología y Salud Pública (CIBERESP) (06/02/0073), FIS 08-1555 and Marato TV3 (051210) (Spain), La Fondation de France, no. 1999 0084 71 (M. Maynadié), Compagnia di San Paolo di Torino, Programma Oncologia 2001 (P.C.), the Health Research Board, Ireland, and the Ministry of Health of the Czech Republic, MZ0 MOU 2005 (L.F.).
Author information
Authors and Affiliations
Contributions
Design and interpretation of overall study: C.F.S., E.H., K.M.B., M.T.S., P.M.B. Primary data analysis: L.C., E.H. Drafting of manuscript: L.C., C.F.S. Critical revision of manuscript: E.H., K.M.B., A.B.-W., B.A., C.M. Vajdic., S.C., M.T.S., P.M.B. Study design, genotyping and statistical analysis of individual studies: SF1, SF1B and SF2: L.C., E.H., J.R., N.K.A., L.A., E.A.H., M.T.S., P.M.B., C.F.S. SCALE: K.E.S., J.L., H.-O.A., H.D., H.H., H.-Q.L., K.H., M. Melbye, E.T.C., B.G. NCI-SEER: N.R., W.C., S.D., P.H., L.M.M., M.S., S.S.W., S.C., J.R.C. NSW: B.A., A.K., S.M., M.P.P., C.M. Vajdic. Yale: P. Boyle, Q.L., S.H.Z., Y.Z., T.Z. EpiLymph: A.N., N.B., Y.B., P. Boffetta, P. Brennan, K.B., P.C., L.F., M. Maynadié, S.de.S., A.S. BC: A.B.-W., J.J.S. Mayo-GEC: S.L.S., S.J.A., T.G.C., M.G., N.J.C., N.E.C., J.R.C., J.M.C., L.R.G., C.A.H., N.E.K., M.C.L., J.F.L., G.E.M., K.G.R., L.Z.R., L.G.S., S.S.S., C.M. Vachon, J.B.W. All authors contributed to the final manuscript and approved its content.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–11, Supplementary Figures 1–5 and Supplementary Methods (PDF 1404 kb)
Rights and permissions
About this article
Cite this article
Conde, L., Halperin, E., Akers, N. et al. Genome-wide association study of follicular lymphoma identifies a risk locus at 6p21.32. Nat Genet 42, 661–664 (2010). https://doi.org/10.1038/ng.626
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.626
This article is cited by
-
Implementation of individualised polygenic risk score analysis: a test case of a family of four
BMC Medical Genomics (2022)
-
Distinct germline genetic susceptibility profiles identified for common non-Hodgkin lymphoma subtypes
Leukemia (2022)
-
Cancer drivers and clonal dynamics in acute lymphoblastic leukaemia subtypes
Blood Cancer Journal (2021)
-
Population-based epidemiological data of follicular lymphoma in Poland: 15 years of observation
Scientific Reports (2020)
-
Infectious mononucleosis, immune genotypes, and non-Hodgkin lymphoma (NHL): an InterLymph Consortium study
Cancer Causes & Control (2020)