Abstract
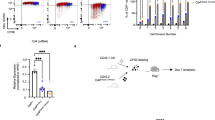
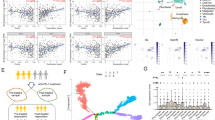
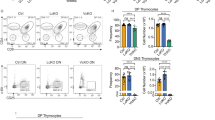
The process of thymocyte development culminates in the maturation of helper (CD4+) and cytotoxic (CD8+) T cells from their common precursors, the CD4+CD8+ double-positive cells1,2. A crucial step during lineage specification is the termination of expression of either the CD4 or the CD8 coreceptor3,4. A silencer element within the first intron of the CD4 gene is sufficient for CD4 transcriptional repression in cells of the cytotoxic lineage, as well as in thymocytes at earlier stages of differentiation5,6. Here we show that the function of the CD4 silencer is required only at distinct stages of development. Its deletion before the initiation of lineage specification resulted in CD4 derepression throughout thymocyte differentiation. By contrast, once cells committed to the cytotoxic CD8+ lineage, the CD4 locus remained silent through subsequent mitoses, even when the silencer element was excised. The epigenetic inheritance of the silenced CD4 locus was not affected by the inhibition of DNA methylation or histone deacetylation, and may thus involve other mechanisms that ensure a stable state of gene expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
von Boehmer, H. T-cell lineage fate: instructed by receptor signals? Curr. Biol. 10, R642–645 (2000).
Berg, L.J. & Kang, J. Molecular determinants of TCR expression and selection. Curr. Opin. Immunol. 13, 232–241 (2001).
Kisielow, P. & von Boehmer, H. Development and selection of T cells: facts and puzzles. Adv. Immunol. 58, 87–209 (1995).
Ellmeier, W., Sawada, S. & Littman, D.R. The regulation of CD4 and CD8 coreceptor gene expression during T cell development. Annu. Rev. Immunol. 17, 523–554 (1999).
Sawada, S., Scarborough, J.D., Killeen, N. & Littman, D.R. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell 77, 917–929 (1994).
Siu, G., Wurster, A.L., Duncan, D.D., Soliman, T.M. & Hedrick, S.M. A transcriptional silencer controls the developmental expression of the CD4 gene. EMBO J. 13, 3570–3579 (1994).
Weiss, A. & Littman, D.R. Signal transduction by lymphocyte antigen receptors. Cell 76, 263–274 (1994).
Basson, M.A. & Zamoyska, R. The CD4/CD8 lineage decision: integration of signalling pathways. Immunol. Today 21, 509–514 (2000).
Marrack, P. & Kappler, J. Positive selection of thymocytes bearing αβ T cell receptors. Curr. Opin. Immunol. 9, 250–255 (1997).
Maldonado, E., Hampsey, M. & Reinberg, D. Repression: targeting the heart of the matter. Cell 99, 455–458 (1999).
Cavalli, G. & Paro, R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell 93, 505–518 (1998).
Grunstein, M. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell 93, 325–328 (1998).
Wolfer, A. et al. Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8T cell development. Nature Immunol. 2, 235–241 (2001).
Ellmeier, W., Sunshine, M.J., Losos, K., Hatam, F. & Littman, D.R. An enhancer that directs lineage-specific expression of CD8 in positively selected thymocytes and mature T cells. Immunity 7, 537–547 (1997).
Suzuki, H., Punt, J.A., Granger, L.G. & Singer, A. Asymmetric signaling requirements for thymocyte commitment to the CD4+ versus CD8+ T cell lineages: a new perspective on thymic commitment and selection. Immunity 2, 413–425 (1995).
Wolffe, A.P. & Matzke, M.A. Epigenetics: regulation through repression. Science 286, 481–486 (1999).
Bird, A.P. & Wolffe, A.P. Methylation-induced repression—belts, braces, and chromatin. Cell 99, 451–454 (1999).
Grunstein, M. Histone acetylation in chromatin structure and transcription. Nature 389, 349–352 (1997).
Bell, A.C. & Felsenfeld, G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405, 482–485 (2000).
Hark, A.T. et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405, 486–489 (2000).
Panning, B. & Jaenisch, R. RNA and the epigenetic regulation of X chromosome inactivation. Cell 93, 305–308 (1998).
Clerc, P. & Avner, P. Genetics. Reprogramming X inactivation. Science 290, 1518–1519 (2000).
Cameron, E.E., Bachman, K.E., Myohanen, S., Herman, J.G. & Baylin, S.B. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nature Genet. 21, 103–107 (1999).
Jones, P.L. et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature Genet. 19, 187–191 (1998).
Csankovszki, G., Nagy, A. & Jaenisch, R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J. Cell Biol. 153, 773–784 (2001).
Imai, S., Armstrong, C.M., Kaeberlein, M. & Guarente, L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 (2000).
Moazed, D. Enzymatic activities of Sir2 and chromatin silencing. Curr. Opin. Cell Biol. 13, 232–238 (2001).
Gu, H., Zou, Y.R. & Rajewsky, K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell 73, 1155–1164 (1993).
DeKoter, R.P., Walsh, J.C. & Singh, H. PU.1 regulates both cytokine-dependent proliferation and differentiation of granulocyte/macrophage progenitors. EMBO J. 17, 4456–4468 (1998).
Kinsella, T.M. & Nolan, G.P. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene. Ther. 7, 1405–1413 (1996).
Van Parijs, L. et al. Uncoupling IL-2 signals that regulate T cell proliferation, survival, and Fas-mediated activation-induced cell death. Immunity 11, 281–288 (1999).
Matzinger, P. The JAM test. A simple assay for DNA fragmentation and cell death. J. Immunol. Methods 145, 185–192 (1991).
Clark, S.J., Harrison, J., Paul, C.L. & Frommer, M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22, 2990–2997 (1994).
Acknowledgements
We thank I. Riviere for her participation in the early phase of this project, J. Hirst for cell sorting, H. Singh for the MSCV–EGFP vector and H. Gu and D.S. Kwon for critical reading of the manuscript. Y.-R.Z. was the recipient of a postdoctoral fellowship from the Irvington Institute and D.R.L. is an Investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Rights and permissions
About this article
Cite this article
Zou, YR., Sunshine, MJ., Taniuchi, I. et al. Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nat Genet 29, 332–336 (2001). https://doi.org/10.1038/ng750
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng750
This article is cited by
-
CD4 expression in effector T cells depends on DNA demethylation over a developmentally established stimulus-responsive element
Nature Communications (2022)
-
Tcf1 and Lef1 provide constant supervision to mature CD8+ T cell identity and function by organizing genomic architecture
Nature Communications (2021)
-
Systematic identification of silencers in human cells
Nature Genetics (2020)
-
Runx-dependent and silencer-independent repression of a maturation enhancer in the Cd4 gene
Nature Communications (2018)
-
Stage-specific epigenetic regulation of CD4 expression by coordinated enhancer elements during T cell development
Nature Communications (2018)