Abstract
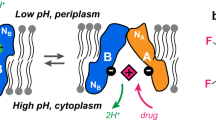
Secondary active transport proteins play a central role in conferring bacterial multidrug resistance. In this work, we investigated the proton-coupled transport mechanism for the Escherichia coli drug efflux pump EmrE using NMR spectroscopy. Our results show that the global conformational motions necessary for transport are modulated in an allosteric fashion by the protonation state of a membrane-embedded glutamate residue. These observations directly correlate with the resistance phenotype for wild-type EmrE and the E14D mutant as a function of pH. Furthermore, our results support a model in which the pH gradient across the inner membrane of E. coli may be used on a mechanistic level to shift the equilibrium of the transporter in favor of an inward-open resting conformation poised for drug binding.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Higgins, C.F. Multiple molecular mechanisms for multidrug resistance transporters. Nature 446, 749–757 (2007).
Bay, D.C., Rommens, K.L. & Turner, R.J. Small multidrug resistance proteins: a multidrug transporter family that continues to grow. Biochim. Biophys. Acta 1778, 1814–1838 (2008).
Rapp, M., Seppälä, S., Granseth, E. & von Heijne, G. Emulating membrane protein evolution by rational design. Science 315, 1282–1284 (2007).
Schuldiner, S. in Membrane Transport Mechanism (ed. Krämer, R.Z. & Ziegler, C.) 233–248 (Springer, Berlin, 2014).
Fleishman, S.J. et al. Quasi-symmetry in the cryo-EM structure of EmrE provides the key to modeling its transmembrane domain. J. Mol. Biol. 364, 54–67 (2006).
Ubarretxena-Belandia, I., Baldwin, J.M., Schuldiner, S. & Tate, C.G. Three-dimensional structure of the bacterial multidrug transporter EmrE shows it is an asymmetric homodimer. EMBO J. 22, 6175–6181 (2003).
Chen, Y.J. et al. X-ray structure of EmrE supports dual topology model. Proc. Natl. Acad. Sci. USA 104, 18999–19004 (2007).
Muth, T.R. & Schuldiner, S. A membrane-embedded glutamate is required for ligand binding to the multidrug transporter EmrE. EMBO J. 19, 234–240 (2000).
Cho, M.K., Gayen, A., Banigan, J.R., Leninger, M. & Traaseth, N.J. Intrinsic conformational plasticity of native EmrE provides a pathway for multidrug resistance. J. Am. Chem. Soc. 136, 8072–8080 (2014).
Gayen, A., Banigan, J.R. & Traaseth, N.J. Ligand-induced conformational changes of the multidrug resistance transporter EmrE probed by oriented solid-state NMR spectroscopy. Angew. Chem. Int. Edn Engl. 52, 10321–10324 (2013).
Morrison, E.A. et al. Antiparallel EmrE exports drugs by exchanging between asymmetric structures. Nature 481, 45–50 (2012).
Schanda, P. & Brutscher, B. Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of seconds. J. Am. Chem. Soc. 127, 8014–8015 (2005).
Adam, Y., Tayer, N., Rotem, D., Schreiber, G. & Schuldiner, S. The fast release of sticky protons: kinetics of substrate binding and proton release in a multidrug transporter. Proc. Natl. Acad. Sci. USA 104, 17989–17994 (2007).
Yerushalmi, H. & Schuldiner, S. An essential glutamyl residue in EmrE, a multidrug antiporter from Escherichia coli. J. Biol. Chem. 275, 5264–5269 (2000).
Wilks, J.C. & Slonczewski, J.L. pH of the cytoplasm and periplasm of Escherichia coli: rapid measurement by green fluorescent protein fluorimetry. J. Bacteriol. 189, 5601–5607 (2007).
Jardetzky, O. Simple allosteric model for membrane pumps. Nature 211, 969–970 (1966).
Wang, J. et al. Imaging membrane protein helical wheels. J. Magn. Reson. 144, 162–167 (2000).
Marassi, F.M. & Opella, S.J. A solid-state NMR index of helical membrane protein structure and topology. J. Magn. Reson. 144, 150–155 (2000).
Dutta, S., Morrison, E.A. & Henzler-Wildman, K.A. Blocking dynamics of the SMR transporter EmrE impairs efflux activity. Biophys. J. 107, 613–620 (2014).
deAzevedo, E.R., Bonagamba, T.J. & Schmidt-Rohr, K. Pure-exchange solid-state NMR. J. Magn. Reson. 142, 86–96 (2000).
Jencks, W.P. Utilization of binding energy and coupling rules for active transport and other coupled vectorial processes. Methods Enzymol. 171, 145–164 (1989).
Poulsen, B.E., Rath, A. & Deber, C.M. The assembly motif of a bacterial small multidrug resistance protein. J. Biol. Chem. 284, 9870–9875 (2009).
Amadi, S.T., Koteiche, H.A., Mishra, S. & McHaourab, H.S. Structure, dynamics, and substrate-induced conformational changes of the multidrug transporter EmrE in liposomes. J. Biol. Chem. 285, 26710–26718 (2010).
Mordoch, S.S., Granot, D., Lebendiker, M. & Schuldiner, S. Scanning cysteine accessibility of EmrE, an H+-coupled multidrug transporter from Escherichia coli, reveals a hydrophobic pathway for solutes. J. Biol. Chem. 274, 19480–19486 (1999).
Brill, S., Sade-Falk, O., Elbaz-Alon, Y. & Schuldiner, S. Specificity determinants in small multidrug transporters. J. Mol. Biol. 427, 468–477 (2015).
Banigan, J.R., Gayen, A., Cho, M.K. & Traaseth, N.J. A structured loop modulates coupling between the substrate-binding and dimerization domains in the multidrug resistance transporter EmrE. J. Biol. Chem. 290, 805–814 (2015).
Rotem, D. & Schuldiner, S. EmrE, a multidrug transporter from Escherichia coli, transports monovalent and divalent substrates with the same stoichiometry. J. Biol. Chem. 279, 48787–48793 (2004).
Brill, S., Falk, O.S. & Schuldiner, S. Transforming a drug/H+ antiporter into a polyamine importer by a single mutation. Proc. Natl. Acad. Sci. USA 109, 16894–16899 (2012).
Son, M.S. et al. Mutagenesis of SugE, a small multidrug resistance protein. Biochem. Biophys. Res. Commun. 312, 914–921 (2003).
Acknowledgements
This work was supported by US National Institutes of Health (NIH) grant R01AI108889 and start-up funds from New York University (NYU) to N.J.T. M.L. acknowledges support from a Margaret Strauss Kramer Fellowship. The NMR data collected with a cryoprobe at NYU was supported by an NIH S10 grant (OD016343). Data collected at the New York Structural Biology Center was made possible by a grant from NYSTAR. The authors thank R. Turner at the University of Calgary for providing wild-type EmrE in the pMS119EH vector, M.-K. Cho for assistance of the initial NMR experiments, A. Sae Her for assistance in protein purification, and D. Buccella at NYU for use of the spectrofluorometer.
Author information
Authors and Affiliations
Contributions
N.J.T. designed the project. A.G. and M.L. carried out the solution NMR, solid-state NMR, fluorescence experiments and ethidium resistance assays. All authors analyzed the data. N.J.T. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–14. (PDF 1934 kb)
Rights and permissions
About this article
Cite this article
Gayen, A., Leninger, M. & Traaseth, N. Protonation of a glutamate residue modulates the dynamics of the drug transporter EmrE. Nat Chem Biol 12, 141–145 (2016). https://doi.org/10.1038/nchembio.1999
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1999
This article is cited by
-
Ion and lipid orchestration of secondary active transport
Nature (2024)
-
Solid-state NMR spectroscopy
Nature Reviews Methods Primers (2021)
-
Structure and dynamics of the drug-bound bacterial transporter EmrE in lipid bilayers
Nature Communications (2021)
-
Multi-receiver solid-state NMR using polarization optimized experiments (POE) at ultrafast magic angle spinning
Journal of Biomolecular NMR (2020)
-
Site-specific resolution of anionic residues in proteins using solid-state NMR spectroscopy
Journal of Biomolecular NMR (2020)