Abstract
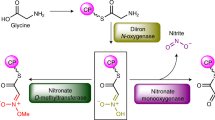
Small molecules containing the N-nitroso group, such as the bacterial natural product streptozotocin, are prominent carcinogens1,2 and important cancer chemotherapeutics3,4. Despite the considerable importance of this functional group to human health, enzymes dedicated to the assembly of the N-nitroso unit have not been identified. Here we show that SznF, a metalloenzyme from the biosynthesis of streptozotocin, catalyses an oxidative rearrangement of the guanidine group of Nω-methyl-l-arginine to generate an N-nitrosourea product. Structural characterization and mutagenesis of SznF reveal two separate active sites that promote distinct steps in this transformation using different iron-containing metallocofactors. This biosynthetic reaction, which has little precedent in enzymology or organic synthesis, expands the catalytic capabilities of non-haem-iron-dependent enzymes to include N–N bond formation. We find that biosynthetic gene clusters that encode SznF homologues are widely distributed among bacteria—including environmental organisms, plant symbionts and human pathogens—which suggests an unexpectedly diverse and uncharacterized microbial reservoir of bioactive N-nitroso metabolites.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The nucleotide sequences for the szn biosynthetic gene cluster and individual genes have been deposited into the NCBI (GenBank accession number for the szn gene cluster: MK303572; Genbank accession numbers for SznA–SznL: MK291255–MK291266). Structural factors and coordinates of SznF have been deposited in the Protein Data Bank (PDB: 6M9R, 6M9S). Additional data that support the conclusions of the paper can be requested from the corresponding authors.
References
Lundberg, J. O., Weitzberg, E. & Gladwin, M. T. The nitrate–nitrite–nitric oxide pathway in physiology and therapeutics. Nat. Rev. Drug Discov. 7, 156–167 (2008).
Lundberg, J. O., Weitzberg, E., Cole, J. A. & Benjamin, N. Nitrate, bacteria and human health. Nat. Rev. Microbiol. 2, 593–602 (2004).
Krug, S. et al. Streptozocin-based chemotherapy in patients with advanced neuroendocrine neoplasms – predictive and prognostic markers for treatment stratification. PLoS ONE 10, e0143822 (2015).
Weiss, R. B. & Issell, B. F. The nitrosoureas: carmustine (BCNU) and lomustine (CCNU). Cancer Treat. Rev. 9, 313–330 (1982).
Vavra, J. J., Deboer, C., Dietz, A., Hanka, L. J. & Sokolski, W. T. Streptozotocin, a new antibacterial antibiotic. Antibiot. Annu. 7, 230–235 (1959).
Wu, K. K. & Huan, Y. Streptozotocin-induced diabetic models in mice and rats. Curr. Protoc. Pharmacol. 40, 5.47.1–5.47.14 (2008).
Pathak, S., Dorfmueller, H. C., Borodkin, V. S. & van Aalten, D. M. Chemical dissection of the link between streptozotocin, O-GlcNAc, and pancreatic cell death. Chem. Biol. 15, 799–807 (2008).
Bolzán, A. D. & Bianchi, M. S. Genotoxicity of streptozotocin. Mutat. Res. 512, 121–134 (2002).
Singaram, S., Lawrence, R. S. & Hornemann, U. Studies on the biosynthesis of the antibiotic streptozotocin (streptozocin) by Streptomyces achromogenes var. streptozoticus. Feeding experiments with carbon-14 and tritium labelled precursors. J. Antibiot. (Tokyo) 32, 379–385 (1979).
Vitturi, D. A. et al. Convergence of biological nitration and nitrosation via symmetrical nitrous anhydride. Nat. Chem. Biol. 11, 504–510 (2015).
Fu, D., Calvo, J. A. & Samson, L. D. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer 12, 104–120 (2012).
Bergy, M. E. et al. Streptozotocin and its production. US Patent 3027300A (1962).
Komor, A. J., Jasniewski, A. J., Que, L. & Lipscomb, J. D. Diiron monooxygenases in natural product biosynthesis. Nat. Prod. Rep. 35, 646–659 (2018).
Lee, J., Simurdiak, M. & Zhao, H. Reconstitution and characterization of aminopyrrolnitrin oxygenase, a Rieske N-oxygenase that catalyzes unusual arylamine oxidation. J. Biol. Chem. 280, 36719–36727 (2005).
Wang, C. et al. Structural determinants for the strict monomethylation activity by Trypanosoma brucei protein arginine methyltransferase 7. Structure 22, 756–768 (2014).
Rui, Z. et al. Microbial biosynthesis of medium-chain 1-alkenes by a nonheme iron oxidase. Proc. Natl Acad. Sci. USA 111, 18237–18242 (2014).
Crane, B. R., Sudhamsu, J. & Patel, B. A. Bacterial nitric oxide synthases. Annu. Rev. Biochem. 79, 445–470 (2010).
Liebschner, D. et al. Polder maps: improving OMIT maps by excluding bulk solvent. Acta Crystallogr. D 73, 148–157 (2017).
Poulos, T. L. Heme enzyme structure and function. Chem. Rev. 114, 3919–3962 (2014).
Rajakovich, L. J. Exploring the functional and mechanistic diversity of diiron oxidases and oxygenases. PhD thesis, The Pennsylvania State Univ. (2017).
Schwarzenbacher, R. et al. Structure of the Chlamydia protein CADD reveals a redox enzyme that modulates host cell apoptosis. J. Biol. Chem. 279, 29320–29324 (2004).
Merkx, M. et al. Dioxygen activation and methane hydroxylation by soluble methane monooxygenase: a tale of two irons and three proteins. Angew. Chem. Int. Ed. 40, 2782–2807 (2001).
Kal, S. & Que, L. Dioxygen activation by nonheme iron enzymes with the 2-His-1-carboxylate facial triad that generate high-valent oxoiron oxidants. J. Biol. Inorg. Chem. 22, 339–365 (2017).
Caranto, J. D., Vilbert, A. C. & Lancaster, K. M. Nitrosomonas europaea cytochrome P460 is a direct link between nitrification and nitrous oxide emission. Proc. Natl Acad. Sci. USA 113, 14704–14709 (2016).
Moënne-Loccoz, P. Spectroscopic characterization of heme iron–nitrosyl species and their role in NO reductase mechanisms in diiron proteins. Nat. Prod. Rep. 24, 610–620 (2007).
Hermeau, R. et al. Gramibactin is a bacterial siderophore with a diazeniumdiolate ligand system. Nat. Chem. Biol. 14, 841–843 (2018).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Darling, A. C., Mau, B., Blattner, F. R. & Perna, N. T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403 (2004).
Söding, J., Biegert, A. & Lupas, A. N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 (2005).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Eswar, N. et al. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics 15, 5.6.1–5.6.30 (2006).
Gust, B., Challis, G. L., Fowler, K., Kieser, T. & Chater, K. F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl Acad. Sci. USA 100, 1541–1546 (2003).
Xia, Y. & Zweier, J. L. Direct measurement of nitric oxide generation from nitric oxide synthase. Proc. Natl Acad. Sci. USA 94, 12705–12710 (1997).
Kunkel, T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl Acad. Sci. USA 82, 488–492 (1985).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Markowitz, V. M. et al. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 40, D115–D122 (2012).
Winter, J. M., Jansma, A. L., Handel, T. M. & Moore, B. S. Formation of the pyridazine natural product azamerone by biosynthetic rearrangement of an aryl diazoketone. Angew. Chem. Int. Ed. 48, 767–770 (2009).
Wang, K. A. et al. Glutamic acid is a carrier for hydrazine during the biosyntheses of fosfazinomycin and kinamycin. Nat. Commun. 9, 3687 (2018).
Forist, A. A. Spectrophotometric determination of streptozotocin. Anal. Chem. 36, 1338–1339 (1964).
Sugishima, M. et al. Crystal structure of dimeric heme oxygenase-2 from Synechocystis sp. PCC 6803 in complex with heme. Biochemistry 44, 4257–4266 (2005).
Adams, N. E. et al. Promiscuous and adaptable enzymes fill “holes” in the tetrahydrofolate pathway in Chlamydia species. MBio 5, e01378-14 (2014).
Pei, J., Kim, B. H., Tang, M. & Grishin, N. V. PROMALS web server for accurate multiple protein sequence alignments. Nucleic Acids Res. 35, W649–W652 (2007).
Magnusson, O. T. et al. Quinone biogenesis: Structure and mechanism of PqqC, the final catalyst in the production of pyrroloquinoline quinone. Proc. Natl Acad. Sci. USA 101, 7913–7918 (2004).
Toms, A. V., Haas, A. L., Park, J. H., Begley, T. P. & Ealick, S. E. Structural characterization of the regulatory proteins TenA and TenI from Bacillus subtilis and identification of TenA as a thiaminase II. Biochemistry 44, 2319–2329 (2005).
Acknowledgements
We thank J. Wang for assistance with LC–MS method development and analysis, M. Wilson for assistance with chemical synthesis and NMR characterization, J. Bergman for assistance with crystallography experiments, and L. Rajakovich for assistance with crystallography data analysis and reading the manuscript. We thank J. M. Bollinger Jr. and C. Krebs for discussions of proposed mechanisms and W. Zhang for providing E. coli WM6026. We acknowledge support from the National Institutes of Health (DP2 GM105434 to E.P.B. and GM119707 to A.K.B.), a Cottrell Scholar Award (to E.P.B.), a Camille Dreyfus Teacher-Scholar Award (to E.P.B.), the Searle Scholars Program (to A.K.B.), and Harvard University. GM/CA@APS has been funded in whole or in part with Federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357. The Eiger 16M detector was funded by an NIH–Office of Research Infrastructure Programs, High-End Instrumentation Grant (1S10OD012289-01A1). Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (grant 085P1000817).
Author information
Authors and Affiliations
Contributions
T.L.N. and E.P.B. initiated the study. T.L.N. performed bioinformatics analyses and located the gene cluster, carried out the in vivo gene knockout and feeding experiments, biochemical characterization of SznE and SznF, chemical syntheses of substrates and standards, liquid chromatography and mass spectrometry analyses, and site-directed mutagenesis experiments. A.K.B. and A.J.M designed the structure determination component of the study. R.R and A.J.M. performed all crystallography experiments with assistance from A.K.B in data analysis. All authors analysed and discussed the results and prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Inorganic nitrogen sources are not precursors to SZN.
a, The results of previous feeding experiments suggested that d-glucosamine, l-citrulline or l-arginine, and l-methionine-derived SAM would be building blocks for SZN biosynthesis9. b, Mass spectra of culture extracts in which 15N-nitrate, 15N-nitrite or 15N-ammonium chloride were fed to S. achromogenes var. streptozoticus NRRL 2697. The expected masses ([M + H]+) for SZN, [15N]SZN, [15N2]SZN and [15N3]SZN are 266.0983, 267.0953, 268.0923 and 269.0894, respectively. These results contrast sharply with the strong labelling (>75%) observed in studies of pathways that use nitrite for diazo biosynthesis37,38.
Extended Data Fig. 2 l-NMA is an on-pathway intermediate in SZN biosynthesis.
a, Comparative genomic analysis of S. achromogenes var. achromogenes NRRL B-2120, a non-producer of SZN and S. achromogenes var. streptozoticus NRRL 2697 using Mauve28. The szn gene cluster is coloured in red. Gene annotations are tabulated in Supplementary Table 1. b, Multiple sequence alignment of SznE with structurally characterized protein arginine methyl transferases (PRMT) from eukaryotes. Conserved residues involved in binding l-arginine are marked with an asterisk. c, Overlay of a homology model of SznE (green) with the crystal structure of PRMT7 from T. brucei (PDB: 4M37) (orange). The highlighted carboxylate residues are involved in binding of the basic guanidine group15. d, SDS–PAGE of purified SznE. The expected molecular weight is 40 kDa. Ladder = Precision Plus Protein All Blue Standards (BioRad). e, Mass spectra of SZN produced when feeding S. achromogenes var. streptozoticus NRRL with d3-l-NMA. The expected masses [M − H2O + H]+ for SZN and d3-SZN are 248.0883 and 251.1060, respectively. f, LC–MS traces demonstrating the restoration of SZN production by the ΔsznE mutant upon chemical complementation with l-NMA. The EICs are generated within a 5 p.p.m. window.
Extended Data Fig. 3 The N-nitrosourea of SZN is derived from an intact guanidine group of l-arginine.
a, The mass spectrum of SZN [M − H2O + H]+ when 1 mM of [15N413C6]l-arginine was added to the fermentation culture. To determine whether the labelled SZN was a single isotopologue or a mixture, degradation (b) and MS/MS (c) experiments were performed. b, Exposure of SZN to ultraviolet light generated a one-carbon- and a one-nitrogen-labelled cyclic urea that was previously reported to be a denitrosated SZN product, indicating that the distal nitroso nitrogen is labelled9. c, MS/MS fragmentation of SZN revealed a one-carbon-labelled cyclic carbamate fragment, indicating that both of the N-nitroso nitrogens are labelled.
Extended Data Fig. 4 Analysis of metabolite production by szn mutants.
Insertions of the antibiotic cassette into each of the szn biosynthetic genes were confirmed by PCR. Culture supernatant extracts from each mutant were analysed with LC–HRMS. Extracted ion chromatograms for the amino acids ([M + H]+) were generated with a 5 p.p.m. window.
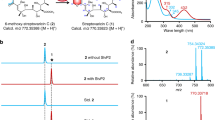
Extended Data Fig. 5 SznF generates an N-nitrosourea-containing amino acid.
a, SDS–PAGE of purified SznF and SznF–SznG complex. The molecular weights of SznF and SznG are 54 kDa and 13 kDa, respectively. Ladder = Precision Plus Protein All Blue Standards (BioRad). b, Nitrite and nitric oxide were detected when Feii–SznF and l-NMA were incubated together. Nitrite was detected with the Griess reagent, and absorbance was measured at 548 nm39. Data are mean ± s.d. of three replicates. NO was trapped with MGD and analysed by EPR spectroscopy at room temperature33. Sodium 2-(N,N-diethylamino)-diazenolate-2-oxide (DEANO) was used as a positive control for NO detection. Assays using [guanidino-15N2]l-NMA as a substrate revealed changes in hyperfine splitting by EPR spectroscopy, indicating that NO is derived from the terminal guanidine group of l-NMA. We propose that the NO detected is derived from the degradation of 3 or is generated as part of the N–N bond-formation step. c, Comparison of retention times and MS/MS fragmentation patterns of 1, 2, Fmoc-3 and 4 generated in SznF assay mixtures with the corresponding synthetic standards. NMR characterization and synthetic procedures are reported in Supplementary Information.
Extended Data Fig. 6 SznF is an iron-dependent monooxygenase.
a, When 1 mM l-arginine, 80 μM SznF, 20 μM PMS and 5 mM NADH were incubated at room temperature for 1 h, only trace amounts of a mass corresponding to l-hydroxyarginine (EIC ([M − H]−) = 189.0993) were observed. No masses corresponding to l-hydroxycitrulline (EIC ([M − H]−) = 190.0883), l-dihydroxyarginine (EIC ([M − H]−) = 205.0942) or the l-nitrosocitrulline (EIC([M − H])− = 203.0786) were observed. b, The [M − H]− mass spectrum of 3 generated when [15N413C6]l-NMA and unlabelled l-NMA were mixed in the same SznF reaction mixture. c, Testing the metal dependence of SznF. 80 μM of apo-SznF was incubated with 200 μM of various divalent metals, 20 μM PMS, 1 mM l-NMA and 5 mM NADH for 1 h at room temperature. The EIC traces were generated with a 5 p.p.m. window. d, Oxygen was rapidly consumed in the presence of l-NMA and SznF as measured by an optode. SznF(E281A), which lacks a key predicted iron-binding residue in the central domain, failed to consume oxygen above background. The background consumption of oxygen arises from the non-enzymatic reduction of PMS by NADH. e, Incubating 18O2, 1 mM l-HMA (1) and 80 μM SznF at room temperature for 1 h resulted in labelling of two of the N-nitrosourea oxygens. MS/MS analysis revealed retention of the Nδ-OH (data not shown). f, Addition of H218O to an SznF assay mixture did not label the N-nitrosourea group. The expected [M − H]− masses for Fmoc-3, [18O]Fmoc-3, [18O2]Fmoc-3 and [18O3]Fmoc-3 are 455.1572, 457.1615, 459.1657 and 461.1700, respectively. g, Addition of catalase or superoxide dismutase to the assay mixtures did not affect SznF-catalysed N-oxygenation as measured by the Griess assay. Data are mean ± s.d. of three replicates.
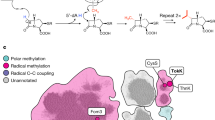
Extended Data Fig. 7 Topology diagram and iron anomalous difference maps for putative SznF catalytic domains.
a, A diagram of the secondary structures found in the N-terminal domain (blue), central helical bundle domain (orange), and C-terminal cupin domain (green) of SznF. Feii ligands and proposed active-site residues are indicated as black dots. Disordered regions are shown as dashed lines. The cupin Feii-binding site is depicted as a circle. b, An Fe anomalous difference map (orange mesh, contoured at 5.0σ) is shown for the fully occupied mononuclear histidine-coordinated Feii site (orange sphere) in the cupin domain. Selected amino acids are shown in stick format. c, The central domain contains a partially occupied (around 50%) iron-binding site in selected crystals, with a smaller peak in the iron anomalous difference map (orange mesh, contoured at 3.0σ).
Extended Data Fig. 8 Comparison of the SznF central domain to haem and diiron structural homologues.
a, SznF contains a large cavity (grey surface, 1.9 Å probe radius) in the middle of its central helical bundle domain (orange). Additionally, most of the secondary structures in this domain contain loop disruptions and disordered regions, suggesting considerable refolding upon binding or release of the l-NMA substrate and/or assembly of the iron-based cofactor. b, The central domain of SznF is similar in topology to haem oxygenase (HO), compared here to HO-2 from Synechocystis sp. PCC 6803 (PDB: 1WOW)40. SznF contains an open pocket near the haem-binding site in HO-2 but lacks conserved cofactor ligation and hydrogen-bonding motifs (d). c, SznF instead more closely resembles a C. trachomatis dinuclear iron protein in this structural superfamily (CADD)21 implicated in the biosynthesis of para-aminobenzoic acid41. SznF conserves all of the metal-binding residues but fails to stably incorporate iron in this domain in the current preparations. All three systems share a propensity for distorted secondary-structure motifs that perhaps enable complex formation with large and polar substrates for oxidative transformations. d, A structure-based sequence alignment42 of six HO-like enzymes in selected regions relevant to substrate–cofactor interaction and catalysis. SznF conserves all six histidine and carboxylate residues used to coordinate a dinuclear iron cluster in the active form of fatty acid oxidative decarboxylase UndA (PDB: 4WWZ)16 and in the uncharacterized CADD protein (PDB: 1RCW)21. Before our discovery and characterization of SznF, UndA was the only HO-like enzyme with a defined substrate and activity. The published crystal structure of UndA contains only a single iron ion and a mechanism was initially proposed using a mononuclear cofactor (located in site 1)16. However, recent spectroscopic studies20 show that this enzyme uses a dinuclear non-haem iron cofactor and corresponding alternative reaction pathway. So far, the dinuclear form of UndA has remained refractory to crystallographic characterization owing to a propensity for disorder in the helix containing the site 2 metal ion ligands. As in SznF, mutagenesis of any of the six predicted ligands to the recently characterized dinuclear site in UndA completely abolishes activity20. As a consequence, we propose that all HO-like non-haem-iron proteins (including SznF) assemble a multinuclear cofactor but require a second protein or other factor to stabilize the active form in high yield and at high concentration. e, Comparative views of the cofactor site and/or substrate binding site in (left to right, top to bottom) SznF, C. trachomatis CADD, Pseudomonas fluorescens UndA, Klebsiella pneumoniae pyrroloquinoline quinone (PQQ) synthase PqqC (PDB: 1OTW)43, Synechocystis sp. PCC 6803 haem oxygenase (HO) 2 (PDB: 1WOW)40, and Bacillus subtilis thiamin synthase TenA (PDB: 1YAK)44. Substrates, products and selected side chains are shown in stick format. Iron ions and water molecules are shown as orange and red spheres, respectively. f, Additional mutation of the predicted iron-binding residues in the SznF central bundle helix domain abolished N-oxygenation activity. Assay mixtures contained 1 mM l-NMA, 80 μM SznF or variant, 20 μM PMS and 5 mM NADH and were incubated at room temperature for 1 h. The EIC traces were generated with a 5 p.p.m. window using the [M − H]− masses.
Extended Data Fig. 9 The binding mode of 1 in the SznF C-terminal cupin domain and assays with a constitutional isomer suggest that the Nδ–OH group is critical for the oxidative rearrangement.
a, An extended water-mediated hydrogen-bonding network tethers the non-metabolizable ligand 1 (green sticks, black lines) in the active site via its Me–Nω, Nδ–O(H), and backbone amine and carboxylate functional groups. Selected SznF amino acids are shown in stick format in a and in grey lines in b. Hydrogen-bonding and ionic interactions are shown as grey (a) or blue (b) dashed lines. Analysis of the network suggests a mechanism for deprotonation of 1 Me-Nω via Y459 and E98. The cupin active site also contains an open hydrophobic pocket near the unmethylated Nω position. Apart from the aforementioned Y459 interaction, there are no hydrogen bonds between the substrate functional groups directly involved in the rearrangement reaction and residues in the active site. b, Ligand-interaction map showing Feii-coordination interactions (distances in Å) and hydrogen-bonding interactions with selected side chains and water molecules. c, Use of the substrate analogue Nω-hydroxy-Nω-methyl-l-arginine (5) at 1 mM final concentration with 80 μM SznF, 20 μM PMS and 5 mM NADH resulted in production of only trace amounts of 3 after incubation for 1 h at room temperature. No [M − H]− masses corresponding to 2, 3 without an Nδ–OH (EIC = 217.0942), 4, or 4 without an Nδ–OH (EIC = 188.1041) were observed. d, The reaction catalysed by the cupin domain does not require an external reductant. The conversion from 2 to 3 proceeded when 1 mM of 2 was incubated with 80 μM SznF without NADH at room temperature for 1 h. The EIC traces were generated with a 5 p.p.m. window.
Extended Data Fig. 10 Distribution of SznF homologues in microbial genomes.
a, Maximum-likelihood phylogenetic tree inferred from 50 replicates showing the relationship between selected SznF homologues containing both the central domain and cupin domain (NCBI Non-Redundant Protein Sequences database, 2018) (E <1 × 10−50). The branch corresponding to S. achromogenes SznF is highlighted by a single asterisk. Bootstrap confidence values of >50 are indicated by black circles on the nodes. The amino acid sequence of UndA is used as an outgroup (highlighted as ∗∗). The sequences used to generate this tree are tabulated in Supplementary Table 2. b, Distribution of 352 SznF homologues that contain both a central domain and a cupin domain in different bacterial genera (IMG/JGI ‘all isolates’ database, 2018) (E <1 × 10−5). See also Supplementary Table 3. c, Selected biosynthetic gene clusters that encode homologues of SznF.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Figures 1–11, Supplementary Tables 1–7, Supplementary Discussion and Supplementary References.
Rights and permissions
About this article
Cite this article
Ng, T.L., Rohac, R., Mitchell, A.J. et al. An N-nitrosating metalloenzyme constructs the pharmacophore of streptozotocin. Nature 566, 94–99 (2019). https://doi.org/10.1038/s41586-019-0894-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-0894-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.