Abstract
Fungi-induced plant diseases affect global food security and plant ecology. The biotrophic fungus Ustilago maydis causes smut disease in maize (Zea mays) plants by secreting numerous virulence effectors that reprogram plant metabolism and immune responses1,2. The secreted fungal chorismate mutase Cmu1 presumably affects biosynthesis of the plant immune signal salicylic acid by channelling chorismate into the phenylpropanoid pathway3. Here we show that one of the 20 maize-encoded kiwellins (ZmKWL1) specifically blocks the catalytic activity of Cmu1. ZmKWL1 hinders substrate access to the active site of Cmu1 through intimate interactions involving structural features that are specific to fungal Cmu1 orthologues. Phylogenetic analysis suggests that plant kiwellins have a versatile scaffold that can specifically counteract pathogen effectors such as Cmu1. We reveal the biological activity of a member of the kiwellin family, a widely conserved group of proteins that have previously been recognized only as important human allergens.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Coordinates and structure factors were deposited at the Protein Data Bank (PDB) under the accession codes 6FPF, 6FPG, 6HJW and 6H3P for Cmu1, the Cmu1–ZmKWL1 complex, ZmCM1 and ZmCM2, respectively. Extended Data Figs. 3, 5, 7 have associated source data. The data that support the findings of this study are available from the corresponding authors upon reasonable request.
References
Lanver, D. et al. Ustilago maydis effectors and their impact on virulence. Nat. Rev. Microbiol. 15, 409–421 (2017).
Matei, A. & Doehlemann, G. Cell biology of corn smut disease — Ustilago maydis as a model for biotrophic interactions. Curr. Opin. Microbiol. 34, 60–66 (2016).
Djamei, A. et al. Metabolic priming by a secreted fungal effector. Nature 478, 395–398 (2011).
Kumar, D. Salicylic acid signaling in disease resistance. Plant Sci. 228, 127–134 (2014).
Bekal, S., Niblack, T. L. & Lambert, K. N. A chorismate mutase from the soybean cyst nematode Heterodera glycines shows polymorphisms that correlate with virulence. Mol. Plant Microbe Interact. 16, 439–446 (2003).
Doyle, E. A. & Lambert, K. N. Meloidogyne javanica chorismate mutase 1 alters plant cell development. Mol. Plant Microbe Interact. 16, 123–131 (2003).
Liu, T. et al. Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 5, 4686 (2014).
Xue, Y., Lipscomb, W. N., Graf, R., Schnappauf, G. & Braus, G. The crystal structure of allosteric chorismate mutase at 2.2-A resolution. Proc. Natl Acad. Sci. USA 91, 10814–10818 (1994).
Sträter, N., Schnappauf, G., Braus, G. & Lipscomb, W. N. Mechanisms of catalysis and allosteric regulation of yeast chorismate mutase from crystal structures. Structure 5, 1437–1452 (1997).
Lanver, D. et al. The biotrophic development of Ustilago maydis studied by RNA-seq analysis. Plant Cell 30, 300–323 (2018).
Mei, Y., Zhang, C., Kernodle, B. M., Hill, J. H. & Whitham, S. A. A Foxtail mosaic virus vector for virus-induced gene silencing in maize. Plant Physiol. 171, 760–772 (2016).
Bölker, M., Genin, S., Lehmler, C. & Kahmann, R. Genetic regulation of mating and dimorphism in Ustilago Maydis. Can. J. Bot. 73, 320–325 (1995).
Di Stasio, M., Brefort, T., Mendoza-Mendoza, A., Münch, K. & Kahmann, R. The dual specificity phosphatase Rok1 negatively regulates mating and pathogenicity in Ustilago maydis. Mol. Microbiol. 73, 73–88 (2009).
Franco, F. P. et al. The sugarcane defense protein SUGARWIN2 causes cell death in Colletotrichum falcatum but not in non-pathogenic fungi. PLoS ONE 9, e91159 (2014).
Wang, N., Xiao, B. & Xiong, L. Identification of a cluster of PR4-like genes involved in stress responses in rice. J. Plant Physiol. 168, 2212–2224 (2011).
Offermann, L. R. et al. Elusive structural, functional, and immunological features of Act d 5, the green kiwifruit kiwellin. J. Agric. Food Chem. 63, 6567–6576 (2015).
Hamiaux, C. et al. Crystal structure of kiwellin, a major cell-wall protein from kiwifruit. J. Struct. Biol. 187, 276–281 (2014).
Draffehn, A. M. et al. Comparative transcript profiling by SuperSAGE identifies novel candidate genes for controlling potato quantitative resistance to late blight not compromised by late maturity. Front. Plant Sci. 4, 423 (2013).
Mosquera, T. et al. Targeted and untargeted approaches unravel novel candidate genes and diagnostic SNPs for quantitative resistance of the potato (Solanum tuberosum L.) to Phytophthora infestans causing the late blight disease. PLoS ONE 11, e0156254 (2016).
Quintana-Camargo, M. et al. Identification of genes differentially expressed in husk tomato (Physalis philadelphica) in response to whitefly (Trialeurodes vaporariorum) infestation. Acta Physiol. Plant. 37, 29 (2015).
Koncz, C. & Schell, J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Genet. Genomics 204, 383–396 (1986).
Kämper, J. et al. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444, 97–101 (2006).
Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular cloning: a laboratory manual 2nd edn (Cold Spring Harbor Laboratory, New York, 1989).
Müller, P., Weinzierl, G., Brachmann, A., Feldbrügge, M. & Kahmann, R. Mating and pathogenic development of the Smut fungus Ustilago maydis are regulated by one mitogen-activated protein kinase cascade. Eukaryot. Cell 2, 1187–1199 (2003).
Brachmann, A., Weinzierl, G., Kämper, J. & Kahmann, R. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42, 1047–1063 (2001).
Loubradou, G., Brachmann, A., Feldbrügge, M. & Kahmann, R. A homologue of the transcriptional repressor Ssn6p antagonizes cAMP signalling in Ustilago maydis. Mol. Microbiol. 40, 719–730 (2001).
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D 69, 1204–1214 (2013).
Pannu, N. S. et al. Recent advances in the CRANK software suite for experimental phasing. Acta Crystallogr. D 67, 331–337 (2011).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Sasso, S., Ramakrishnan, C., Gamper, M., Hilvert, D. & Kast, P. Characterization of the secreted chorismate mutase from the pathogen Mycobacterium tuberculosis. FEBS J. 272, 375–389 (2005).
Holliday, R. Molecular aspects of genetic exchange and gene conversion. Genetics 78, 273–287 (1974).
Wales, T. E., Fadgen, K. E., Gerhardt, G. C. & Engen, J. R. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal. Chem. 80, 6815–6820 (2008).
Geromanos, S. J. et al. The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics 9, 1683–1695 (2009).
Li, G. Z. et al. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics 9, 1696–1719 (2009).
Schäper, S. et al. AraC-like transcriptional activator CuxR binds c-di-GMP by a PilZ-like mechanism to regulate extracellular polysaccharide production. Proc. Natl Acad. Sci. USA 114, E4822–E4831 (2017).
Schuhmacher, J. S. et al. MinD-like ATPase FlhG effects location and number of bacterial flagella during C-ring assembly. Proc. Natl Acad. Sci. USA 112, 3092–3097 (2015).
Steinchen, W. et al. Catalytic mechanism and allosteric regulation of an oligomeric (p)ppGpp synthetase by an alarmone. Proc. Natl Acad. Sci. USA 112, 13348–13353 (2015).
Rost, B. Twilight zone of protein sequence alignments. Protein Eng. 12, 85–94 (1999).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M. & Barton, G. J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
Ronquist, F. et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165 (2011).
Edgar, R. C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 (2004).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Li, W. et al., The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 43, W580–W584 (2015).
Holm, L. & Rosenstrom, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010).
Huet, J. et al. High-resolution structure of a papaya plant-defense barwin-like protein solved by in-house sulfur-SAD phasing. Acta Crystallogr. D 69, 2017–2026 (2013).
Karniel, A. et al. Co-translational folding intermediate dictates membrane targeting of the signal recognition particle receptor. J. Mol. Biol. 430, 1607–1620 (2018).
Acknowledgements
We thank S. A. Whitham for the FoMV silencing system, V. Vincon for help in scoring the VIGS experiment and C. Pellegrin for interpreting RNA sequencing data. G.B. acknowledges support by the Deutsche Forschungsgemeinschaft (DFG) through the DFG-core facility for interactions, dynamics and macromolecular assembly structure and the Collaborative Research Council SFB987. F.A. and X.H. were supported by the Peter and Traudl Engelhorn foundation and the China Scholarship Council (CSC), respectively. R.K. acknowledges generous support by the Max Planck Society. We thank the European Synchrotron Radiation Facility (ESRF) for support.
Reviewer information
Nature thanks N. Talbot and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
X.H. conducted in vivo experiments, western blotting and enzyme kinetic analysis. T.G. identified proteins by mass spectrometry. F.A. and J.S. determined all crystal structures. W.S. analysed the ZmKWL1/Cmu1 complex by hydrogen–deuterium exchange mass spectrometry. F.A. and L.B. performed pull-down analysis. P.I.G. performed isothermal titration calorimetry analysis. A.D. provided the relevant material. S.A.R. performed phylogenetic analysis. R.K., S.R. and G.B. planned experiments, analysed data and supervised the project. R.K. and G.B. wrote the manuscript. All authors read and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Comparison of Cmu1 and Aro7p.
a, Sequence alignment of Aro7p and Cmu1. Conserved active site residues are coloured in red. b, Side-by-side view of the active sites of Aro7p and Cmu1 shows that all residues that are critical for substrate binding and catalysis are conserved. To prepare the figure, the structures of Aro7p (left; PDB: 3CSM9 and Cmu1 (right; this study) were superimposed. Aro7p is in complex with an endo-oxabicyclic transition state analogue inhibitor (TSA). c, Left, cysteine residues 203 and 289 of Cmu1 form a disulfide bridge linking helix α5 and the C terminus. Right, the 2Fo − Fc electron density map at a 1.6σ cut-off around the disulfide bridge bond. It is of note that both cysteines are highly conserved among Cmu1 homologues from plant-pathogenic fungi and are not found in housekeeping chorismate mutases, such as Aro7p (see a).
Extended Data Fig. 2 Comparison of chorismate mutases of the maize plant.
a, Sequence alignment of Zea mays chorismate mutases. The alignment was generated with MUSCLE47,48,49, with residues colour-coded according to their chemical properties, and the table was generated in Chimera33. b, c, Crystal structures of ZmCM1 from the chloroplast (b) and ZmCM2 from the cytoplasm (c) of Z. mays (this study). A dashed line divides the homodimer of each structure. Both structures show the conserved chorismate mutase fold, including an allosteric loop. d, Comparison between Cmu1, CM1 from A. thaliana (At) (PDB: 4PPU), ZmCM1 and ZmCM2. The ELR and allosteric loop are highlighted in red.
Extended Data Fig. 3 Z. mays KWL1 interacts with U. maydis Cmu1 in vivo and in vitro and is involved in infection of maize by U. maydis.
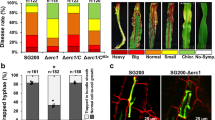
a, Mass spectrometry unambiguously identified the hypothetical maize protein with the annotation GRMZM2G073114 (ZmKWL1). The protein was identified by several unique spectra of peptides (indicated in yellow) covering 44 per cent of the total sequence. b, Relative expression levels of 20 maize kiwellin genes during biotrophic development of U. maydis were revealed by RNA sequencing analysis of RNA samples collected from mock (top) and FB1 × FB2 (bottom) infected maize plants (dataset available via ref. 10 and GEO accession number GSE103876). The vertical axis indicates normalized counts from DESeq2 analysis. The horizontal axis indicates the infection stages: 0.5, 1, 2, 4, 6, 8 and 12 days post infection (dpi). Data represent mean ± s.d. of n = 3 biological replicates. Colour codes for different genes are indicated in the legend. c, Six-day-old maize seedlings were rub-inoculated with virus-sap containing pFoMV-V and pFoMV-ZmKWL1181–480, respectively. Twenty-seven leaves from three biological replicates were analysed six days after viral inoculation and expression of ZmKWL1 was analysed separately in each leaf by qPCR using the maize gene GAPDH, which encodes glyceraldehyde-3-phosphate-dehydrogenase, as internal reference. Boxplot representation indicates that expression levels of ZmKWL1 in pFoMV-ZmKWL1181–480 infected maize leaves were reduced compared to expression levels in pFoMV-V infected plants, indicative of silencing. A total of 27 leaves from n = 3 biological replicates were assessed. The centre line of the box plot denotes the median, lower and upper box hinges represent first and third quartiles, respectively, and whiskers indicate the maximum and minimum values. P value was determined by unpaired two-sided Student’s t-test. *P = 0.0202. d, Maize plants silenced for ZmKWL1 via infection with pFoMV-ZmKWL1181–480 (left) and non-silenced plants infected with pFoMV-V (right) were infected with CL13 strain. Representative leaves with heaviest U. maydis infection symptoms were imaged 12 days after CL13 infection. Representative experiments are shown. The experiment was repeated independently twice with similar results. e, Supernatant of AB33-p123-Potef-Cmu1-HA3 containing Cmu1-HA3 was incubated with Ni-NTA agarose and plant lysates of N. benthamiana leaves infiltrated with GV3101 transformed with empty vector pEZRK-LCY or pEZRK-ZmKWL1-His6. ZmKWL1 and Cmu1 were detected by western blot using His antibody (top) and HA antibody (bottom), respectively. Representative images are displayed. The experiment was repeated independently once with similar results.
Extended Data Fig. 4 The ZmKWL1 interaction site at Cmu1 is specific for secreted chorismate mutases from plant-pathogenic fungi.
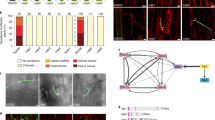
a, Left, representative regions of Cmu1 lining the interaction interface are indicated in the crystal structure of the Cmu1–ZmKWL1 complex. Right, deuterium uptake plots of representative regions of Cmu1 alone (red) and the Cmu1–ZmKWL1 complex (blue). Data represent mean ± s.d. of n = 3 technical replicates. b, Amino acid sequences of chorismate mutases from the plant-pathogenic fungi Ustilago maydis (UmCmu1), Melanopsichium pennsylvanicum (MpCmu1), Sporisorium scitamineum (SsCmu1), Sporisorium reilianum (SrCmu1), Ustilago hordei (UhCmu1), Ustilago bromivora (UbCmu1) and chorismate mutases from the non-pathogenic species Saccharomyces cerevisiae (ScAro7p), Zea mays (ZmCM1 and ZmCM2) and A. thaliana (AtCM2) were aligned with MUSCLE47,48,49. Conserved amino acid residues are colour-coded according to their polarity and charge. The secondary structure of UmCmu1 (this study) is displayed above the alignment. The disulfide-bond-forming cysteine residues Cys203 and Cys289 (numbering according to UmCmu1) found in plant-pathogenic fungi are indicated. Chorismate mutases critically differ in the region between helices α2 and α3. In plant-pathogenic fungi, the additional helix α2a and an extended loop region (ELR, numbering according to UmCmu1) are present.
Extended Data Fig. 5 Prerequisites of Cmu1 for the Cmu1–KWL1 interaction and localization of ZmKWL1.
a, ZmKWL1 is unable to interact with the Cmu1-ΔELR variant. Supernatant of AB33-p123-Potef-Cmu1-HA3 or AB33-p123-Potef-Cmu1-ΔELR-HA3 containing Cmu1-HA3 or Cmu1-ΔELR-HA3, was incubated with Ni-NTA agarose and plant lysate of N. benthamiana leaves infiltrated with GV3101 transformed with pEZRK-ZmKWL1-His6. After co-immunoprecipitation, ZmKWL1 and Cmu1 were detected by western blot using antibodies against His6 (ZmKWL1, top) or HA3 (Cmu1, bottom), respectively. Representative images are displayed. The experiment was repeated independently once with similar results. b, ZmKWL1 inhibits the chorismate mutase activity of Cmu1 but not of Cmu1 that lacks the ELR (Cmu1-ΔELR). Chorismate mutase activity was determined with the chorismate mutase activity assay. Data represent mean ± s.d. of n = 3 technical replicates. c, To investigate the localization of ZmKWL1, we transiently expressed ZmKWL1-sfGFP fusion proteins with (ZmKWL1-sfGFP) and without signal peptide (ZmKWL1ΔSP-sfGFP) using A. tumefaciens for delivery. ZmKWL1ΔSP-sfGFP mainly accumulated in the cytosol and the nucleus. After plasmolysis with 1 M NaCl, the fluorescence signal pattern remained unchanged. By contrast, ZmKWL1-sfGFP with its native signal peptide showed peripheral localization and the signal accumulated in the enlarged apoplastic space after plasmolysis. This observation demonstrates the secretion of ZmKWL1 into the apoplast. The dashed blue line marks the plant cell wall and the dashed yellow line marks the plant plasma membrane. Arrows mark the enlarged apoplast in which the secreted ZmKWL1-sfGFP protein accumulates. Scale bars, 25 µm. Representative images are displayed. The experiment was repeated independently once with similar results.
Extended Data Fig. 6 Amino acid sequence alignment of different kiwellin paralogs from Z. mays.
Amino acid sequences from Z. mays kiwellins were aligned with MUSCLE47,48,49. The secondary structure of ZmKWL1 (GRMZM2G073114) is displayed according to the crystal structure. Conserved residues are colour-coded according to their polarity and charge. Kiwellin homologues are highly conserved in their β-barrel domains. Five kiwellins show similarity to the β1/β2 region of ZmKWL1. Four kiwellins carry C-terminal extensions.
Extended Data Fig. 7 Interaction of different maize kiwellins with Cmu1 and similarities among kiwellins, barwin-like proteins and cerato-platanin.
a, ZmKWL4 (GRMZM2G432697) migrates with an apparent molecular mass of 15 kDa, which corresponds to a monomer (calculated molecular mass: 17 kDa), whereas Cmu1 migrates with the apparent molecular mass of a dimer. When ZmKWL4 and Cmu1were mixed in a 2:1 ratio, no complex formation was observed. b, ZmKWL12 (GRMZM2G429533) migrates with an apparent molecular mass of 15 kDa, which corresponds to a monomer (calculated molecular mass: 17 kDa), whereas Cmu1 migrates with the apparent molecular mass of a dimer. When ZmKWL12 and Cmu1 were mixed in a 2:1 ratio, no complex formation was observed. c, ZmKwl6 (GRMZM2G331599) migrates with an apparent molecular mass of 13 kDa which corresponds to a monomer (calculated molecular mass: 14 kDa), while Cmu1 migrates with the apparent molecular mass of a dimer. When ZmKwl6 and Cmu1were mixed in a 2:1 ratio, again no complex formation was observed. In a–c, the insets show a Coomassie-stained SDS–PAGE of the peak fraction. Each experiment was repeated twice with similar results. d, Activity of Cmu1 is inhibited by ZmKWL1 but not by ZmKWL4, ZmKWL6 or ZmKWL12. Data represent mean ± s.d. of n = 3 technical replicates. e, The crystal structures of two kiwellin proteins from Actinidia chinensis (PDB: 4PMK17) and Actinidia deliciosa (AdKWL, PDB: 4X9U16) superimpose very well with the structure of ZmKWL1 with root mean square deviations (r.m.s.d.) of 0.42 Å and 0.40 Å over 119 Cα-atoms and 110 Cα-atoms, respectively (alignment not shown). Search with the DALI-server50 revealed members of the barwin and CP-proteins as structurally similar to ZmKWL1. The structure of a barwin-like protein from Carica papaya (CpBarwin, PDB: 4JP651) superposes well with ZmKWL1 (r.m.s.d.: 0.92 Å over all Cα-atoms). The β-barrel domain of ZmKWL1 and the barwin-like protein formed by six β-strands can be perfectly superposed. The β1/β2-domain of ZmKWL1, however, is absent in barwin-like proteins. Moreover, several loops critical for the recognition of Cmu1 by ZmKWL1 are considerably reduced in length in the barwin-like proteins or completely absent. The structure of a cerato-platanin protein from Moniliophthora perniciosa (MpCP2, PDB: 3SUK) also shares the β-barrel with the kiwellins and barwin-like proteins and aligns well with ZmKWL1 (r.m.s.d.: 1.11 Å over all Cα-atoms). Our structural comparison of the barwin-like proteins with the cerato-platanin shows that both proteins are highly similar. This suggests to us that barwin-like proteins and cerato-platanins are, in structural terms, members of the same protein family. All structures are shown as cartoons and are rainbow-coloured from their N- to their C termini. f, g, Topology of ZmKWL1 (f) and a Barwin-like protein (g). The structure of both proteins is coloured in rainbow from blue (N-terminal) to red (C-terminal). A yellow circle indicates individual cysteines, and disulfide bond numbers (1–5, as above) are indicated.
Supplementary information
Supplementary Data
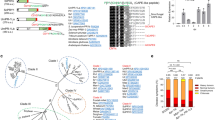
This dataset contains the phylogenetic inference of the kiwellin protein family in plants and fungi. Bayesian inference tree, the numbers at the nodes represent posterior probabilities (see supplemental Methods for details). Sequences of asterids are shown in red, rosids in blue, monocots in green, fungi in grey and all other sequences (non-seed plants, stem angiosperms and eudicots, gymnosperms) in black. Most paralog acquisitions appear to be lineage-specific
Supplementary Information
This file contains uncropped gel data scans
Rights and permissions
About this article
Cite this article
Han, X., Altegoer, F., Steinchen, W. et al. A kiwellin disarms the metabolic activity of a secreted fungal virulence factor. Nature 565, 650–653 (2019). https://doi.org/10.1038/s41586-018-0857-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0857-9
This article is cited by
-
Intrinsically Disordered Kiwellin Protein-Like Effectors Target Plant Chloroplasts and are Extensively Present in Rust Fungi
Molecular Biotechnology (2024)
-
A cell surface-exposed protein complex with an essential virulence function in Ustilago maydis
Nature Microbiology (2021)
-
Physiological and histopathological assessments of the susceptibility of different tomato (Solanum lycopersicum) cultivars to early blight disease
European Journal of Plant Pathology (2021)
-
CdbA is a DNA-binding protein and c-di-GMP receptor important for nucleoid organization and segregation in Myxococcus xanthus
Nature Communications (2020)
-
IM30 IDPs form a membrane-protective carpet upon super-complex disassembly
Communications Biology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.