Abstract
Most adult carcinomas develop from noninvasive precursor lesions, a progression that is supported by genetic analysis. However, the evolutionary and genetic relationships among co-existing lesions are unclear. Here we analysed the somatic variants of pancreatic cancers and precursor lesions sampled from distinct regions of the same pancreas. After inferring evolutionary relationships, we found that the ancestral cell had initiated and clonally expanded to form one or more lesions, and that subsequent driver gene mutations eventually led to invasive pancreatic cancer. We estimate that this multi-step progression generally spans many years. These new data reframe the step-wise progression model of pancreatic cancer by illustrating that independent, high-grade pancreatic precursor lesions observed in a single pancreas often represent a single neoplasm that has colonized the ductal system, accumulating spatial and genetic divergence over time.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Sequence data have been deposited at the European Genome-phenome Archive (EGA; https://www.ebi.ac.uk/ega/), which is hosted by the European Bioinformatics Institute (EBI) and the Centre for Genomic Regulation (CRG), under accession number EGAS00001002778. Source data are provided for Fig. 3b and Extended Data Figs. 1, 7, 8. All other relevant data are included within the manuscript or are available upon request from the corresponding author.
References
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013).
Vogelstein, B. & Kinzler, K. W. The path to cancer —three strikes and you’re out. N. Engl. J. Med. 373, 1895–1898 (2015).
Basturk, O. et al. A revised classification system and recommendations from the Baltimore consensus meeting for neoplastic precursor lesions in the pancreas. Am. J. Surg. Pathol. 39, 1730–1741 (2015).
Hruban, R. H., Goggins, M., Parsons, J. & Kern, S. E. Progression model for pancreatic cancer. Clin. Cancer Res. 6, 2969–2972 (2000).
van Heek, N. T. et al. Telomere shortening is nearly universal in pancreatic intraepithelial neoplasia. Am. J. Pathol. 161, 1541–1547 (2002).
Kanda, M. et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology 142, 730–733.e9 (2012).
Makohon-Moore, A. & Iacobuzio-Donahue, C. A. Pancreatic cancer biology and genetics from an evolutionary perspective. Nat. Rev. Cancer 16, 553–565 (2016).
Murphy, S. J. et al. Genetic alterations associated with progression from pancreatic intraepithelial neoplasia to invasive pancreatic tumor. Gastroenterology 145, 1098–1109.e1 (2013).
Hosoda, W. et al. Genetic analyses of isolated high-grade pancreatic intraepithelial neoplasia (HG-PanIN) reveal paucity of alterations in TP53 and SMAD4. J. Pathol. 242, 16–23 (2017).
Fearon, E. R. & Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 61, 759–767 (1990).
Matsuda, Y. et al. The prevalence and clinicopathological characteristics of high-grade pancreatic intraepithelial neoplasia: autopsy study evaluating the entire pancreatic parenchyma. Pancreas 46, 658–664 (2017).
Yamasaki, S., Suda, K., Nobukawa, B. & Sonoue, H. Intraductal spread of pancreatic cancer. Clinicopathologic study of 54 pancreatectomized patients. Pancreatology 2, 407–412 (2002).
Kleeff, J. et al. Pancreatic cancer. Nat. Rev. Dis. Primers 2, 16022 (2016).
Notta, F. et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature 538, 378–382 (2016).
Roberts, N. J. et al. Whole genome sequencing defines the genetic heterogeneity of familial pancreatic cancer. Cancer Discov. 6, 166–175 (2016).
Jones, S. et al. Comparative lesion sequencing provides insights into tumor evolution. Proc. Natl Acad. Sci. USA 105, 4283–4288 (2008).
Biankin, A. V. et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405 (2012).
Waddell, N. et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518, 495–501 (2015).
Witkiewicz, A. K. et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 6, 6744 (2015).
Bailey, P. et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52 (2016).
Reiter, J. G. et al. Reconstructing metastatic seeding patterns of human cancers. Nat. Commun. 8, 14114 (2017).
Makohon-Moore, A. P. et al. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat. Genet. 49, 358–366 (2017).
Sanli, O. et al. Bladder cancer. Nat. Rev. Dis. Primers 3, 17022 (2017).
Pea, A. et al. Targeted DNA sequencing reveals patterns of local progression in the pancreatic remnant following resection of intraductal papillary mucinous neoplasm (IPMN) of the pancreas. Ann. Surg. 266, 133–141 (2017).
Roerink, S. F. et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature 556, 457–462 (2018).
Yachida, S. et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117 (2010).
Tomasetti, C., Vogelstein, B. & Parmigiani, G. Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proc. Natl Acad. Sci. USA 110, 1999–2004 (2013).
Klein, W. M., Hruban, R. H., Klein-Szanto, A. J. P. & Wilentz, R. E. Direct correlation between proliferative activity and dysplasia in pancreatic intraepithelial neoplasia (PanIN): additional evidence for a recently proposed model of progression. Mod. Pathol. 15, 441–447 (2002).
Hruban, R. H. et al. An illustrated consensus on the classification of pancreatic intraepithelial neoplasia and intraductal papillary mucinous neoplasms. Am. J. Surg. Pathol. 28, 977–987 (2004).
Jones, S. et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806 (2008).
Reiter, J. G. & Iacobuzio-Donahue, C. A. Pancreatic cancer: Pancreatic carcinogenesis—several small steps or one giant leap? Nat. Rev. Gastroenterol. Hepatol. 14, 7–8 (2016).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Mose, L. E., Wilkerson, M. D., Hayes, D. N., Perou, C. M. & Parker, J. S. ABRA: improved coding indel detection via assembly-based realignment. Bioinformatics 30, 2813–2815 (2014).
Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013).
Tokheim, C. J., Papadopoulos, N., Kinzler, K. W., Vogelstein, B. & Karchin, R. Evaluating the evaluation of cancer driver genes. Proc. Natl Acad. Sci. USA 113, 14330–14335 (2016).
Davoli, T. et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155, 948–962 (2013).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
Chang, M. T. et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat. Biotechnol. 34, 155–163 (2016).
Cancer Genome Atlas Research Network Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 32, 185–203.e13 (2017).
Shen, R. & Seshan, V. E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 44, e131 (2016).
Rausch, T. et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28, i333–i339 (2012).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Alexandrov, L. B., Nik-Zainal, S., Wedge, D. C., Campbell, P. J. & Stratton, M. R. Deciphering signatures of mutational processes operative in human cancer. Cell Reports 3, 246–259 (2013).
Amikura, K., Kobari, M. & Matsuno, S. The time of occurrence of liver metastasis in carcinoma of the pancreas. Int. J. Pancreatol. 17, 139–146 (1995).
Durrett, R. in Branching Process Models of Cancer 1–63 (Springer International Publishing, Switzerland, 2015).
Acknowledgements
Supported by the V Foundation for Cancer Research, NIH grants F31 CA180682, 2T32 CA160001-06 and 5T32 CA067751-13, an Erwin Schrödinger fellowship (Austrian Science Fund FWF J-3996), SPORE grant P50 CA062924, the Michael Rolfe Foundation, The Lustgarten Foundation for Cancer Research, the Sol Goldman Center for Pancreatic Cancer Research, The Virginia and D.K. Ludwig Fund for Cancer Research and D. Troper and S. Wojcicki.
Reviewer information
Nature thanks A. V. Biankin, S. J. Chanock and F. Markowetz for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
A.P.M.-M., K.M., Y.J., N.P., K.K., B.V., and C.I.-D. designed the study; K.M., S.Y., R.H.H., and C.I.-D. selected the samples; S.Y., K.M., R.H.H., D.S.K., and C.I.-D. reviewed pathology; S.Y., K.M. and M.Z. prepared the DNA samples; A.P.M.-M., K.M., M.Z., Y.J., N.P., K.K., B.V., and C.I.-D. performed sequencing, alignment and mutation calling; A.P.M.-M., J.G.R., J.M.G., and M.A. derived the phylogenies; A.P.M.-M., J.G.R., J.M.G., M.A., and L.S. analysed the structural variants; J.G.R., J.M.G., and M.A.N. performed mathematical modelling; C.S. illustrated the spatial evolution of the lesions; and all authors wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
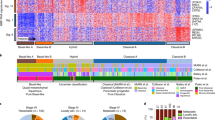
Extended Data Fig. 1 Mutation counts and features of samples.
a, Number of somatic mutations detected per sample with clinical features of each patient. The y-axis shows mutation counts and the x-axis shows patient ID. FS Del, frameshift deletion; FS Ins, frameshift insertion; IF Del, in-frame deletion; IF Ins, in-frame insertion; TSS, transcription start site. b, c, Box and whisker plots comparing number of somatic SNVs and CNAs between PanINs and PDACs. Yellow, PanINs; green, PDACs. Horizontal line indicates median, whiskers indicate minimum and maximum, and box indicates quartiles. n = 12 independent PanIN lesions, n = 8 PDACs. b, Number of SNVs and indels in PanINs and PDACs. c, Number of CNAs in PanINs and PDACs (hisens results only).
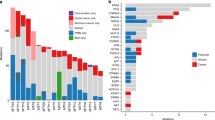
Extended Data Fig. 2 Allelic CNAs across all patient samples.
CNAs were inferred using the FACETS algorithm (Supplementary Table 3, FACETS.purity variants shown). Blue, putative losses; red, putative gains.
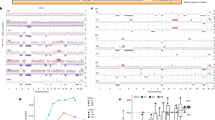
Extended Data Fig. 3 Phylogenetics of PanINs and the matched primary tumour for patients PIN102 and PIN105.
See Supplementary Table 1 for sample identities. Gene names in orange text are SNVs or indels, in blue are copy-number losses, and in red are copy-number gains affecting putative driver genes. The sequencing data for each driver gene variant was manually reviewed to verify phylogenetic position. For each phylogeny, the number of acquired mutations is in black. The branch lengths are proportional to the number of SNVs and indels. The dashed line indicates the branch from the germline to the PDAC and PanIN-A. For the Bayesian heat maps, samples are indicated on each row while variants are represented by each column. The colour of each tile indicates the probability that the variant is present or absent in the corresponding sample. Dark blue, >99.9% probability of being present; dark red, >99.9% probability of being absent. Light blue and red indicate lower probabilities; white tiles indicate approximately 50% probability. a, Phylogenetic tree and Bayesian heat map with each variant for PIN102. b, Phylogenetic tree and Bayesian heat map with each variant for PIN105.
Extended Data Fig. 4 Phylogenetics of PanINs and matched primary tumours for patients PIN101, PIN103, PIN104, and PIN108.
See Supplementary Table 1 for sample identities. Gene names in orange text are SNVs or indels, in blue are copy-number losses, and in red are copy-number gains affecting putative driver genes. The sequencing data for each driver gene variant were manually reviewed to verify phylogenetic position. For each phylogenetic tree, the numbers of acquired mutations are in black. The branch lengths are proportional to the number of SNVs or indels. The dashed lines indicate branches that have been extended to accommodate gene annotation and variant numbers. For the Bayesian heatmaps, samples are indicated on each row and variants are represented by each column. The colour of each tile indicates the probability that the variant is present or absent in the corresponding sample. Dark blue, >99.9% probability of being present; dark red, >99.9% probability of being absent. Light blue and red indicate lower probabilities; white tiles indicate approximately 50% probability. a, PIN101. In manual review of the sequencing data, a read supporting the presence of the KRAS(G12D) variant was detected in both the PDAC and PanIN-A samples and was thus moved to the trunk of the phylogeny despite the overall low coverage of KRAS in PanIN-A. b, PIN103. c, PIN104. The node leading from the first MRCA to the second MRCA has a confidence value of >99%. d, PIN108. The node leading from the first MRCA to the second MRCA has a confidence value of >99%.
Extended Data Fig. 5 Phylogenetics of PanINs and matched primary tumours for patients PIN106 and PIN107.
See Supplementary Table 1 for sample identities. Gene names in orange text are SNVs or indels, in blue are copy-number losses, and in red are copy-number gains affecting putative driver genes. The sequencing data for each driver gene variant were manually reviewed to verify phylogenetic position. For each phylogeny, the numbers of acquired mutations are in black. The branch lengths are proportional to the number of SNVs or indels. The dashed lines indicate branches that have been extended to accommodate gene annotation and variant numbers. For each Bayesian heat map, samples are indicated on each row while variants are represented by each column. The colour of each tile indicates the probability that the variant is present or absent in the corresponding sample. Dark blue, >99.9% probability of being present; dark red, >99.9% probability of being absent. Light blue and red indicate lower probabilities; white tiles indicate approximately 50% probability. a, PIN106. The node leading from the first MRCA to the second MRCA has a confidence value of >99% and the node leading from the second MRCA to the third MRCA has a confidence value of 82%. b, PIN107.
Extended Data Fig. 6 Average signature abundance across samples.
Signature numbers 1–30 from Alexandrov et al.43 are shown on the x-axis with signature abundance averaged across phylogenetic branches shown on the y-axis. Each histogram is coloured by signature identity.
Extended Data Fig. 7 The proportion of mutational signatures from Alexandrov et al.43 estimated in PIN101–PIN104.
Signatures are shown on the x-axis, with the proportion of each signature shown on the y-axis. Each bar is coloured by signature identity. The text on the top of each panel denotes the corresponding phylogenetic branch and the number of mutations acquired along it in parentheses. Error bars depict 90% CIs in the signature proportion estimated by 100 iterations of bootstrap resampling.
Extended Data Fig. 8 The proportion of mutational signatures from Alexandrov et al.43 estimated in PIN105–PIN108.
Signatures are shown on the x-axis, with the proportion of each signature shown on the y-axis. Each bar is coloured by signature identity. The text on the top of each panel denotes the corresponding phylogenetic branch and the number of mutations acquired along it in parentheses. Error bars depict 90% CIs in the signature proportion estimated by 100 iterations of bootstrap resampling.
Supplementary information
Supplementary Table 1
Clinical features of each patient.
Supplementary Table 2
Somatic SNVs/INDELs detected among PanIN and PDAC lesions.
Supplementary Table 3
Putative somatic CNAs among PanIN and PDAC lesions.
Supplementary Table 4
Structural variants in each patient.
Supplementary Table 5
Putative driver gene SNVs/INDELs.
Supplementary Table 6
Jaccard similarity coefficients between all pairs of samples within each case.
Rights and permissions
About this article
Cite this article
Makohon-Moore, A.P., Matsukuma, K., Zhang, M. et al. Precancerous neoplastic cells can move through the pancreatic ductal system. Nature 561, 201–205 (2018). https://doi.org/10.1038/s41586-018-0481-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0481-8
Keywords
This article is cited by
-
Optimization of a mouse model of pancreatic cancer to simulate the human phenotypes of metastasis and cachexia
BMC Cancer (2024)
-
3D genomic mapping reveals multifocality of human pancreatic precancers
Nature (2024)
-
Improving the prognosis of pancreatic cancer: insights from epidemiology, genomic alterations, and therapeutic challenges
Frontiers of Medicine (2023)
-
Integrated clinical and genomic analysis identifies driver events and molecular evolution of colitis-associated cancers
Nature Communications (2023)
-
Evolutionary histories of breast cancer and related clones
Nature (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.