Abstract
Sexual reproduction is restricted to eukaryotic species and involves the fusion of haploid gametes to form a diploid cell that subsequently undergoes meiosis to generate recombinant haploid forms. This process has been extensively studied in the unicellular yeast Saccharomyces cerevisiae, which exhibits separate regulatory control over mating and meiosis. Here we address the mechanism of sexual reproduction in the related hemiascomycete species Candida lusitaniae. We demonstrate that, in contrast to S. cerevisiae, C. lusitaniae exhibits a highly integrated sexual program in which the programs regulating mating and meiosis have fused. Profiling of the C. lusitaniae sexual cycle revealed that gene expression patterns during mating and meiosis were overlapping, indicative of co-regulation. This was particularly evident for genes involved in pheromone MAPK signalling, which were highly induced throughout the sexual cycle of C. lusitaniae. Furthermore, genetic analysis showed that the orthologue of IME2, a ‘diploid-specific’ factor in S. cerevisiae1,2, and STE12, the master regulator of S. cerevisiae mating3,4, were each required for progression through both mating and meiosis in C. lusitaniae. Together, our results establish that sexual reproduction has undergone significant rewiring between S. cerevisiae and C. lusitaniae, and that a concerted sexual cycle operates in C. lusitaniae that is more reminiscent of the distantly related ascomycete, Schizosaccharomyces pombe. We discuss these results in light of the evolution of sexual reproduction in yeast, and propose that regulatory coupling of mating and meiosis has evolved multiple times as an adaptation to promote the haploid lifestyle.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
19 February 2014
The online Abstract was corrected to match the print and PDF versions of this paper.
References
Irniger, S. The Ime2 protein kinase family in fungi: more duties than just meiosis. Mol. Microbiol. 80, 1–13 (2011)
Yoshida, M. et al. Initiation of meiosis and sporulation in Saccharomyces cerevisiae requires a novel protein kinase homologue. Mol. Gen. Genet. 221, 176–186 (1990)
Dolan, J. W., Kirkman, C. & Fields, S. The yeast STE12 protein binds to the DNA sequence mediating pheromone induction. Proc. Natl Acad. Sci. USA 86, 5703–5707 (1989)
Errede, B. & Ammerer, G. STE12, a protein involved in cell-type-specific transcription and signal transduction in yeast, is part of protein-DNA complexes. Genes Dev. 3, 1349–1361 (1989)
Souza, C. A., Silva, C. C. & Ferreira, A. V. Sex in fungi: lessons of gene regulation. Genet. Mol. Res. 2, 136–147 (2003)
van Werven, F. J. & Amon, A. Regulation of entry into gametogenesis. Phil. Trans. R. Soc. Lond. B 366, 3521–3531 (2011)
Govin, J. & Berger, S. L. Genome reprogramming during sporulation. Int. J. Dev. Biol. 53, 425–432 (2009)
Butler, G. et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459, 657–662 (2009)
Reedy, J. L., Floyd, A. M. & Heitman, J. Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex. Curr. Biol. 19, 891–899 (2009)
Gargeya, I. B., Pruitt, W. R., Simmons, R. B., Meyer, S. A. & Ahearn, D. G. Occurrence of Clavispora lusitaniae, the teleomorph of Candida lusitaniae, among clinical isolates. J. Clin. Microbiol. 28, 2224–2227 (1990)
Wong Sak Hoi, J. & Dumas, B. Ste12 and Ste12-like proteins, fungal transcription factors regulating development and pathogenicity. Eukaryot. Cell 9, 480–485 (2010)
Lengeler, K. B. et al. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64, 746–785 (2000)
Butler, G. Fungal sex and pathogenesis. Clin. Microbiol. Rev. 23, 140–159 (2010)
Keeney, S. in Recombination and Meiosis Vol. 2 (eds Egel, R. & Lankenau, D. ) (Springer, 2007)
Klein, F. et al. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98, 91–103 (1999)
Smith, H. E. & Mitchell, A. P. A transcriptional cascade governs entry into meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 9, 2142–2152 (1989)
Chu, S. et al. The transcriptional program of sporulation in budding yeast. Science 282, 699–705 (1998)
Guttmann-Raviv, N., Martin, S. & Kassir, Y. Ime2, a meiosis-specific kinase in yeast, is required for destabilization of its transcriptional activator, Ime1. Mol. Cell. Biol. 22, 2047–2056 (2002)
Holt, L. J., Hutti, J. E., Cantley, L. C. & Morgan, D. O. Evolution of Ime2 phosphorylation sites on Cdk1 substrates provides a mechanism to limit the effects of the phosphatase Cdc14 in meiosis. Mol. Cell 25, 689–702 (2007)
Sopko, R., Raithatha, S. & Stuart, D. Phosphorylation and maximal activity of Saccharomyces cerevisiae meiosis-specific transcription factor Ndt80 is dependent on Ime2. Mol. Cell. Biol. 22, 7024–7040 (2002)
Lin, C. H., Choi, A. & Bennett, R. J. Defining pheromone-receptor signaling in Candida albicans and related asexual Candida species. Mol. Biol. Cell 22, 4918–4930 (2011)
Young, L. Y., Lorenz, M. C. & Heitman, J. A STE12 homolog is required for mating but dispensable for filamentation in Candida lusitaniae. Genetics 155, 17–29 (2000)
Sipiczki, M. Where does fission yeast sit on the tree of life? Genome Biol. 1, reviews1011 (2000)
Sugimoto, A., Iino, Y., Maeda, T., Watanabe, Y. & Yamamoto, M. Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev. 5, 1990–1999 (1991)
Kjaerulff, S., Lautrup-Larsen, I., Truelsen, S., Pedersen, M. & Nielsen, O. Constitutive activation of the fission yeast pheromone-responsive pathway induces ectopic meiosis and reveals Ste11 as a mitogen-activated protein kinase target. Mol. Cell. Biol. 25, 2045–2059 (2005)
Herskowitz, I. A regulatory hierarchy for cell specialization in yeast. Nature 342, 749–757 (1989)
Booth, L. N., Tuch, B. B. & Johnson, A. D. Intercalation of a new tier of transcription regulation into an ancient circuit. Nature 468, 959–963 (2010)
Otto, S. P. The evolutionary consequences of polyploidy. Cell 131, 452–462 (2007)
Otto, S. P. & Gerstein, A. C. The evolution of haploidy and diploidy. Curr. Biol. 18, R1121–R1124 (2008)
Roberts, C. J. et al. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287, 873–880 (2000)
Guthrie, C. & Fink, G. R. Guide to Yeast Genetics and Molecular Biology Vol. 194 (Academic Press, 1991)
Reuß, O., Vik, A., Kolter, R. & Morschhauser, J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341, 119–127 (2004)
Cregg, J. M. et al. in Methods Enzymology Vol. 463 (eds Burgess, R. R. & Deutscher, M. P. ) 169–189 (Elsevier, 2009)
El-Kirat-Chatel, S., Dementhon, K. & Noël, T. A two-step cloning-free PCR-based method for the deletion of genes in the opportunistic pathogenic yeast Candida lusitaniae. Yeast 28, 321–330 (2011)
François, F., Chapeland-Leclerc, F., Villard, J. & Noël, T. Development of an integrative transformation system for the opportunistic pathogenic yeast Candida lusitaniae using URA3 as a selection marker. Yeast 21, 95–106 (2004)
Kooistra, R., Hooykaas, P. J. & Steensma, H. Y. Efficient gene targeting in Kluyveromyces lactis. Yeast 21, 781–792 (2004)
Wach, A., Brachat, A., Pohlmann, R. & Philippsen, P. Dominant marker vectors for selecting yeast mating products. Yeast 52, 595–599 (2008)
Noble, S. M. & Johnson, A. D. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4, 298–309 (2005)
Lin-Cereghino, J. et al. Direct selection of Pichia pastoris expression strains using new G418 resistance vectors. Yeast 25, 293–299 (2008)
Barth, G. & Gaillardin, C. Physiology and genetics of the dimorphic fungus Yarrowia lipolytica. FEMS Microbiol. Rev. 19, 219–237 (1997)
Schiestl, R. H., Manivasakam, P., Woods, R. A. & Gietzt, R. D. Introducing DNA into yeast by transformation. Methods 5, 79–85 (1993)
Lemoine, S., Combes, F., Servant, N. & Le Crom, S. Goulphar: rapid access and expertise for standard two-color microarray normalization methods. BMC Bioinformatics 7, 467 (2006)
Saldanha, A. J. Java Treeview—extensible visualization of microarray data. Bioinformatics 20, 3246–3248 (2004)
de Hoon, M. J. L., Imoto, S., Nolan, J. & Miyano, S. Open source clustering software. Bioinformatics 20, 1453–1454 (2004)
Flores, C. L., Gancedo, C. & Petit, T. Disruption of Yarrowia lipolytica TPS1 gene encoding trehalose-6-P synthase does not affect growth in glucose but impairs growth at high temperature. PLoS ONE 6, e23695 (2011)
Acknowledgements
We thank J. Heitman, C. Lisset-Flores Mauriz, T. Noel, N. Hunter and J. Reedy for gifts of strains and plasmids, and S. Kabrawala and N. Balmuri for help with strain construction. We also thank T. Sorrells, L. Holt and members of the Johnson and Bennett laboratories for comments on the paper, and S. Jones for help with statistical analysis. This work was supported by National Science Foundation Grant MCB1021120 (to R.J.B.), by National Institutes of Health R01 Grant AI081704 (to R.J.B.), by T32GM007601 (to C.M.S.), by F31AI075607 (to R.K.S.), and by an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund (to R.J.B.).
Author information
Authors and Affiliations
Contributions
R.K.S. and C.M.S. constructed strains, analysed phenotypes and performed transcriptional profiling experiments. S.E.T. and R.J.B. constructed strains and analysed phenotypes. R.K.S., C.M.S. and R.J.B. were involved in study design and writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
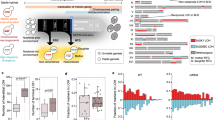
Extended Data Figure 1 Schematic showing induction of pheromone-processing genes (a) and pheromone MAPK genes (b) during both mating and meiosis in C. lusitaniae.
Expression changes highlight genes induced during co-incubation of a and α cells on PDA medium (mating), as well as during growth of diploid a/α cells on PDA medium (induces meiosis and sporulation). In contrast, these genes are mating-specific in the related yeast S. cerevisiae. Scale indicates fold change for gene expression.
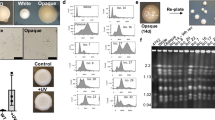
Extended Data Figure 2 Temporal analysis of meiosis in C. lusitaniae.
a, C. lusitaniae a/α cells divide as stable diploid cells on YPD medium. n = 3. b, On PDA medium, a/α cells are induced to enter meiosis and dyad spores (asterisks) begin to appear at 18 h. n = 3. c, Time course of meiosis in wild type, ste12Δ /ste12Δ and STE12-complemented C. lusitaniae strains. A morphology change (polarized growth) is evident in wild-type diploid strains grown on PDA medium starting at 12 h, whereas spore formation is apparent at 18–36 h. In the ste12Δ/ste12Δ mutant, both sporulation and morphology change are absent. d, Global transcriptional profile of meiosis in C. lusitaniae showing induced genes (yellow) and repressed genes (blue). C. lusitaniae diploid a/α cells (RSY432) were grown on PDA (sporulating) medium and expression changes compared to those on YPD (non-sporulating) medium. Genes changing more than four fold in expression are shown. All n values represent number of biological replicates. Scale indicates fold expression changes.
Extended Data Figure 3 Transcriptional profiling of C. lusitaniae wild type and ime2Δ mutants during mating and meiosis.
a, Full profile of wild-type and ime2Δ/ime2Δ strains during meiosis indicates that many transcriptional changes occur in ime2Δ/ime2Δ mutants as in wild-type strains. b, Analysis of cell cycle regulating genes during C. lusitaniae meiosis. Several cell cycle genes are induced in wild-type cells undergoing meiosis but not in ime2Δ/ime2Δ mutants (for example, APC4, CDC3, CDC10 and CDC14). c, Comparative expression of early (IME2, IME4, SPO11), middle (REC102, CDC3, SPS4, SPS1, NDT80, SWM1) and late (DIT1, DIT2) meiosis genes. Expression changes scaled as in Extended Data Fig. 1. d, The mating profiles of wild-type and ime2Δ strains were very similar. e, Comparison of meiosis genes induced in wild type and ime2Δ/ime2Δ mutants.
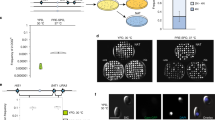
Extended Data Figure 4 Reintegration of IME2 restores mating efficiency.
IME2 was reintegrated at the endogenous locus in an ime2Δ mutant (RSY437). Mating frequency was quantified by monitoring the formation of prototrophic products from auxotrophic parents. WT cross, RSY411 × RSY284, ime2Δ mutant cross, RSY406 × RSY437, IME2 complemented cross, RSY406 × CAY5022. Differences between the WT cross and the ime2Δ mutant cross, and between the IME2 complemented cross and the ime2Δ mutant cross were both significant. n = 9 (3 biological replicates in triplicate), P < 0.001, Kruskal–Wallis test. Data are representative of mean ± s.e.m.
Extended Data Figure 5 Schematic of genetically marked C. lusitaniae strains and control of meiosis by STE12.
a, C. lusitaniae mating experiments were performed between RSY284 (a strain) and RSY411 (α strain) and mating products selected based on auxotrophic makers. b, The a/α diploid strain RSY432 was induced to undergo meiosis on PDA medium and meiotic progeny identified by their red colour on YPD medium (ade− colonies) or by growth on medium containing cycloheximide (CHX-resistant colonies). c, Diploid a/α strains of C. lusitaniae were incubated on PDA medium for 3 days and analysed for the frequency of formation of meiotic progeny. Two independent ste12Δ/ste12Δ mutants were constructed in the a/α background and tested for meiotic progeny using both the CHXR and ADE2 markers. Mutants lacking STE12 were unable to undergo meiosis to produce haploid progeny, while reintegration of STE12 into the mutant background restored meiosis. Differences between both wild-type strains and ste12Δ/ste12Δ mutants, and between ste12Δ/ste12Δ mutants and STE12-complemented strains were significant (n = 3 biological replicates, P < 0.05, student’s t-test, two tailed). Data representative of mean ± s.e.m.
Extended Data Figure 6 Analysis of STE12 function in mating and meiosis in diverse hemiascomycete species.
The STE12 gene was deleted from haploid and diploid strains of K. lactis, P. pastoris and S. cerevisiae. The resultant strains were tested both for mating competency and the formation of meiotic progeny. Whereas deletion of STE12 blocked mating in haploid strains of all three species, loss of STE12 from diploid a/α strains did not have a significant effect on the formation of meiotic progeny. Thus, C. lusitaniae is unique among the hemiascomycete species tested in that STE12 is essential for meiosis only in this species. K. lactis mating and meiosis, n = 2, data combined for 3 independent mutants. P. pastoris mating, n = 3. P. pastoris meiosis n = 4. S. cerevisiae meiosis n = 3. All data presented as mean ± s.e.m. All n values represent number of biological replicates.
Extended Data Figure 7 Rewiring of the genetic programs that control sexual reproduction in hemiascomycetes.
In many hemiascomycete species, including the model yeast S. cerevisiae, mating and meiosis are controlled by two distinct transcriptional programs with STE12 as the master regulator of mating and IME2 as a key regulator of meiosis. However, in the opportunistic pathogen C. lusitaniae, STE12 has retained its role in regulating mating, but has also acquired control over meiosis. Similarly, C. lusitaniae IME2 has a conserved role in regulating mating, but also has a role in promoting mating. The programs controlling these two processes have therefore fused in C. lusitaniae, perhaps to facilitate a transient diploid state and more efficient return to the predominant haploid state.
Supplementary information
Supplementary Information
The online Abstract was corrected to match the print and PDF versions of this paper. (PDF 454 kb)
Rights and permissions
About this article
Cite this article
Sherwood, R., Scaduto, C., Torres, S. et al. Convergent evolution of a fused sexual cycle promotes the haploid lifestyle. Nature 506, 387–390 (2014). https://doi.org/10.1038/nature12891
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12891
This article is cited by
-
Mating type specific transcriptomic response to sex inducing pheromone in the pennate diatom Seminavis robusta
The ISME Journal (2021)
-
DNA damage response of major fungal pathogen Candida glabrata offers clues to explain its genetic diversity
Current Genetics (2021)
-
A ‘parameiosis’ drives depolyploidization and homologous recombination in Candida albicans
Nature Communications (2019)
-
Metschnikowia mating genomics
Antonie van Leeuwenhoek (2018)
-
Regulation of mating type switching by the mating type genes and RME1 in Ogataea polymorpha
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.