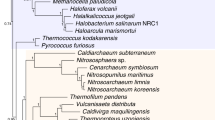
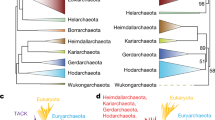
Abstract
The discovery of the Archaea and the proposal of the three-domains ‘universal’ tree, based on ribosomal RNA and core genes mainly involved in protein translation, catalysed new ideas for cellular evolution and eukaryotic origins. However, accumulating evidence suggests that the three-domains tree may be incorrect: evolutionary trees made using newer methods place eukaryotic core genes within the Archaea, supporting hypotheses in which an archaeon participated in eukaryotic origins by founding the host lineage for the mitochondrial endosymbiont. These results provide support for only two primary domains of life—Archaea and Bacteria—because eukaryotes arose through partnership between them.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Woese, C. R. & Fox, G. E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl Acad. Sci. USA 74, 5088–5090 (1977)A landmark paper that, together with ref. 4, reported the discovery of the Archaea and discussed its far-reaching implications for early evolution.
Embley, T. M. & Martin, W. Eukaryotic evolution, changes and challenges. Nature 440, 623–630 (2006)
Woese, C. R. On the evolution of cells. Proc. Natl Acad. Sci. USA 99, 8742–8747 (2002)
Woese, C. R. & Fox, G. E. The concept of cellular evolution. J. Mol. Evol. 10, 1–6 (1977)
Doolittle, W. F. & Brown, J. R. Tempo, mode, the progenote, and the universal root. Proc. Natl Acad. Sci. USA 91, 6721–6728 (1994)
Iwabe, N., Kuma, K., Hasegawa, M., Osawa, S. & Miyata, T. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc. Natl Acad. Sci. USA 86, 9355–9359 (1989)Together with ref. 7 , this paper presented the first evidence for rooting the tree of life on the bacterial stem, but see ref. 5 for a still-relevant discussion of these analyses and other contemporary ideas about early evolution.
Gogarten, J. P. et al. Evolution of the vacuolar H+-ATPase: implications for the origin of eukaryotes. Proc. Natl Acad. Sci. USA 86, 6661–6665 (1989)
Dagan, T., Roettger, M., Bryant, D. & Martin, W. Genome networks root the tree of life between prokaryotic domains. Genome Biol. Evol. 2, 379–392 (2010)
Lake, J. A., Skophammer, R. G., Herbold, C. W. & Servin, J. A. Genome beginnings: rooting the tree of life. Phil. Trans. R. Soc. B 364, 2177–2185 (2009)
Skophammer, R. G., Servin, J. A., Herbold, C. W. & Lake, J. A. Evidence for a gram-positive, eubacterial root of the tree of life. Mol. Biol. Evol. 24, 1761–1768 (2007)
Cavalier-Smith, T. Rooting the tree of life by transition analyses. Biol. Direct 1, 19 (2006)
Cox, C. J., Foster, P. G., Hirt, R. P., Harris, S. R. & Embley, T. M. The archaebacterial origin of eukaryotes. Proc. Natl Acad. Sci. USA 105, 20356–20361 (2008)The first of a series of recent papers demonstrating that analyses of core genes using new phylogenetic models favour the eocyte tree rather than the three-domains tree.
Doolittle, W. F. & Zhaxybayeva, O. in The Prokaryotes: Prokaryotic Biology and Symbiotic Associations (ed. Rosenberg, E. ) (Springer, 2013)A very clear discussion about the issues facing the integration of phylogenetics and classification given the evidence for extensive lateral gene transfer.
Woese, C. R., Kandler, O. & Wheelis, M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl Acad. Sci. USA 87, 4576–4579 (1990)Woese and colleagues present their arguments for the rooted three-domains tree of life.
Madigan, M. T., Martingo, J. M., Stahl, D. A. & Clark, D. P. Brock Biology of Microorganisms 13th edn (Benjamin Cummings, 2010)
Pace, N. R. Time for a change. Nature 441, 289 (2006)
Pace, N. R. Mapping the tree of life: progress and prospects. Microbiol. Mol. Biol. Rev. 73, 565–576 (2009)
Lake, J. A., Henderson, E., Oakes, M. & Clark, M. W. Eocytes: a new ribosome structure indicates a kingdom with a close relationship to eukaryotes. Proc. Natl Acad. Sci. USA 81, 3786–3790 (1984)This paper presents comparisons of ribosomal structure in Bacteria, Archaea and eukaryotes, providing the initial motivation for the eocyte hypothesis.
Gribaldo, S., Poole, A. M., Daubin, V., Forterre, P. & Brochier-Armanet, C. The origin of eukaryotes and their relationship with the Archaea: are we at a phylogenomic impasse? Nature Rev. Microbiol. 8, 743–752 (2010)
Knoll, A. H., Javaux, E. J., Hewitt, D. & Cohen, P. Eukaryotic organisms in Proterozoic oceans. Phil. Trans. R. Soc. B 361, 1023–1038 (2006)
Philippe, H. & Forterre, P. The rooting of the universal tree of life is not reliable. J. Mol. Evol. 49, 509–523 (1999)
Foster, P. G., Cox, C. J. & Embley, T. M. The primary divisions of life: a phylogenomic approach employing composition-heterogeneous methods. Phil. Trans. R. Soc. B 364, 2197–2207 (2009)
Penny, D., McComish, B. J., Charleston, M. A. & Hendy, M. D. Mathematical elegance with biochemical realism: the covarion model of molecular evolution. J. Mol. Evol. 53, 711–723 (2001)
Ho, S. Y. & Jermiin, L. Tracing the decay of the historical signal in biological sequence data. Syst. Biol. 53, 623–637 (2004)
Lartillot, N., Brinkmann, H. & Philippe, H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol. Biol. 7 (suppl. 1). S4 (2007)
Philippe, H. et al. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 9, e1000602 (2011)
Gouy, M. & Li, W. H. Phylogenetic analysis based on rRNA sequences supports the archaebacterial rather than the eocyte tree. Nature 339, 145–147 (1989)
Woese, C. R. Bacterial evolution. Microbiol. Rev. 51, 221–271 (1987)
Olsen, G. J. Earliest phylogenetic branchings: comparing rRNA-based evolutionary trees inferred with various techniques. Cold Spring Harb. Symp. Quant. Biol. 52, 825–837 (1987)
Foster, P. G. & Hickey, D. A. Compositional bias may affect both DNA-based and protein-based phylogenetic reconstructions. J. Mol. Evol. 48, 284–290 (1999)
Foster, P. G. Modeling compositional heterogeneity. Syst. Biol. 53, 485–495 (2004)
Hirt, R. P. et al. Microsporidia are related to Fungi: evidence from the largest subunit of RNA polymerase II and other proteins. Proc. Natl Acad. Sci. USA 96, 580–585 (1999)
Lake, J. A. Reconstructing evolutionary trees from DNA and protein sequences: paralinear distances. Proc. Natl Acad. Sci. USA 91, 1455–1459 (1994)
Yang, Z. & Roberts, D. On the use of nucleic acid sequences to infer early branchings in the tree of life. Mol. Biol. Evol. 12, 451–458 (1995)An important early contribution demonstrating that modelling changing nucleotide composition in RNA sequences from different species supported the eocyte tree.
Felsenstein, J. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 27, 401–410 (1978)
Yang, Z. & Rannala, B. Molecular phylogenetics: principles and practice. Nature Rev. Genet. 13, 303–314 (2012)
Lake, J. A. Origin of the eukaryotic nucleus determined by rate-invariant analysis of rRNA sequences. Nature 331, 184–186 (1988)
Sidow, A. & Wilson, A. C. Compositional statistics: an improvement of evolutionary parsimony and its application to deep branches in the tree of life. J. Mol. Evol. 31, 51–68 (1990)
Tourasse, N. J. & Gouy, M. Accounting for evolutionary rate variation among sequence sites consistently changes universal phylogenies deduced from rRNA and protein-coding genes. Mol. Phylogenet. Evol. 13, 159–168 (1999)
Yutin, N., Makarova, K. S., Mekhedov, S. L., Wolf, Y. I. & Koonin, E. V. The deep archaeal roots of eukaryotes. Mol. Biol. Evol. 25, 1619–1630 (2008)
Harris, J. K., Kelley, S. T., Spiegelman, G. B. & Pace, N. R. The genetic core of the universal ancestor. Genome Res. 13, 407–412 (2003)
Katoh, K., Kuma, K. & Miyata, T. Genetic algorithm-based maximum-likelihood analysis for molecular phylogeny. J. Mol. Evol. 53, 477–484 (2001)
Ciccarelli, F. D. et al. Toward automatic reconstruction of a highly resolved tree of life. Science 311, 1283–1287 (2006)
Lake, J. A. The order of sequence alignment can bias the selection of tree topology. Mol. Biol. Evol. 8, 378–385 (1991)
Brown, J. R., Douady, C. J., Italia, M. J., Marshall, W. E. & Stanhope, M. J. Universal trees based on large combined protein sequence data sets. Nature Genet. 28, 281–285 (2001)
Lartillot, N. & Philippe, H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 21, 1095–1109 (2004)One of the most notable improvements in phylogenetic modelling in the last decade, providing a Bayesian framework for accommodating across-site compositional heterogeneity—a key feature of molecular sequence data.
Guy, L. & Ettema, T. J. The archaeal ‘TACK’ superphylum and the origin of eukaryotes. Trends Microbiol. 19, 580–587 (2011)
Williams, T. A., Foster, P. G., Nye, T. M., Cox, C. J. & Embley, T. M. A congruent phylogenomic signal places eukaryotes within the Archaea. Proc. R. Soc. Lond. B 279, 4870–4879 (2012)
Lasek-Nesselquist, E. & Gogarten, J. P. The effects of model choice and mitigating bias on the ribosomal tree of life. Mol. Phylogenet. Evol. 69, 17–38 (2013)
Pester, M., Schleper, C. & Wagner, M. The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr. Opin. Microbiol. 14, 300–306 (2011)
Lloyd, K. G. et al. Predominant archaea in marine sediments degrade detrital proteins. Nature 496, 215–218 (2013)
Graybeal, A. Is it better to add taxa or characters to a difficult phylogenetic problem? Syst. Biol. 47, 9–17 (1998)
Elkins, J. G. et al. A korarchaeal genome reveals insights into the evolution of the Archaea. Proc. Natl Acad. Sci. USA 105, 8102–8107 (2008)
Brochier-Armanet, C., Boussau, B., Gribaldo, S. & Forterre, P. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nature Rev. Microbiol. 6, 245–252 (2008)
Nunoura, T. et al. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 39, 3204–3223 (2011)
Kelly, S., Wickstead, B. & Gull, K. Archaeal phylogenomics provides evidence in support of a methanogenic origin of the Archaea and a thaumarchaeal origin for the eukaryotes. Proc. R. Soc. Lond. B 278, 1009–1018 (2011)
Ettema, T. J., Lindas, A. C. & Bernander, R. An actin-based cytoskeleton in archaea. Mol. Microbiol. 80, 1052–1061 (2011)
Yutin, N. & Koonin, E. V. Archaeal origin of tubulin. Biol. Direct 7, 10 (2012)
Koonin, E. V., Makarova, K. S. & Elkins, J. G. Orthologs of the small RPB8 subunit of the eukaryotic RNA polymerases are conserved in hyperthermophilic Crenarchaeota and “Korarchaeota”. Biol. Direct 2, 38 (2007)
Csurös, M. & Miklos, I. Streamlining and large ancestral genomes in Archaea inferred with a phylogenetic birth-and-death model. Mol. Biol. Evol. 26, 2087–2095 (2009)
Wolf, Y. I., Makarova, K. S., Yutin, N. & Koonin, E. V. Updated clusters of orthologous genes for Archaea: a complex ancestor of the Archaea and the byways of horizontal gene transfer. Biol. Direct 7, 46 (2012)
Ribeiro, S. & Golding, G. B. The mosaic nature of the eukaryotic nucleus. Mol. Biol. Evol. 15, 779–788 (1998)Together with ref. 63 , this paper presented some of the first tree-based evidence that eukaryotes are genomic chimaeras containing some genes that are most similar to those of Bacteria and others to Archaea.
Rivera, M. C., Jain, R., Moore, J. E. & Lake, J. A. Genomic evidence for two functionally distinct gene classes. Proc. Natl Acad. Sci. USA 95, 6239–6244 (1998)
Esser, C. et al. A genome phylogeny for mitochondria among α-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol. Biol. Evol. 21, 1643–1660 (2004)
Alsmark, C. et al. Patterns of prokaryotic lateral gene transfers affecting parasitic microbial eukaryotes. Genome Biol. 14, R19 (2013)
Cotton, J. A. & McInerney, J. O. Eukaryotic genes of archaebacterial origin are more important than the more numerous eubacterial genes, irrespective of function. Proc. Natl Acad. Sci. USA 107, 17252–17255 (2010)
Dagan, T. & Martin, W. The tree of one percent. Genome Biol. 7, 118 (2006)
Doolittle, W. F. & Bapteste, E. Pattern pluralism and the Tree of Life hypothesis. Proc. Natl Acad. Sci. USA 104, 2043–2049 (2007)
Williams, D. et al. A rooted net of life. Biol. Direct 6, 45 (2011)
Creevey, C. J., Doerks, T., Fitzpatrick, D. A., Raes, J. & Bork, P. Universally distributed single-copy genes indicate a constant rate of horizontal transfer. PLoS ONE 6, e22099 (2011)
Boussau, B. et al. Genome-scale coestimation of species and gene trees. Genome Res. 23, 323–330 (2013)
Szollösi, G. J., Boussau, B., Abby, S. S., Tannier, E. & Daubin, V. Phylogenetic modeling of lateral gene transfer reconstructs the pattern and relative timing of speciations. Proc. Natl Acad. Sci. USA 109, 17513–17518 (2012)
Cohen, O., Gophna, U. & Pupko, T. The complexity hypothesis revisited: connectivity rather than function constitutes a barrier to horizontal gene transfer. Mol. Biol. Evol. 28, 1481–1489 (2011)
Jain, R., Rivera, M. C. & Lake, J. A. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl Acad. Sci. USA 96, 3801–3806 (1999)
Butterfield, N. J. Bangiomorpha pubescens n. gen., n. sp.: implications for the evolution of sex, multicellularity, and the Mesoproterozoic/Neoproterozoic radiation of eukaryotes. Paleobiology 26, 386–404 (2000)
Parfrey, L. W., Lahr, D. J., Knoll, A. H. & Katz, L. A. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc. Natl Acad. Sci. USA 108, 13624–13629 (2011)
Brocks, J. J., Logan, G. A., Buick, R. & Summons, R. E. Archean molecular fossils and the early rise of eukaryotes. Science 285, 1033–1036 (1999)
Rasmussen, B., Fletcher, I. R., Brocks, J. J. & Kilburn, M. R. Reassessing the first appearance of eukaryotes and cyanobacteria. Nature 455, 1101–1104 (2008)
Fischer, W. W. Biogeochemistry: life before the rise of oxygen. Nature 455, 1051–1052 (2008)
Ueno, Y., Yamada, K., Yoshida, N., Maruyama, S. & Isozaki, Y. Evidence from fluid inclusions for microbial methanogenesis in the early Archaean era. Nature 440, 516–519 (2006)
Papineau, D., Walker, J. J., Mojzsis, S. J. & Pace, N. R. Composition and structure of microbial communities from stromatolites of Hamelin Pool in Shark Bay, Western Australia. Appl. Environ. Microbiol. 71, 4822–4832 (2005)
Allwood, A. C. et al. Controls on development and diversity of Early Archean stromatolites. Proc. Natl Acad. Sci. USA 106, 9548–9555 (2009)
Tice, M. M. & Lowe, D. R. Photosynthetic microbial mats in the 3,416-Myr-old ocean. Nature 431, 549–552 (2004)
Schopf, J. W. Fossil evidence of Archaean life. Phil. Trans. R. Soc. B 361, 869–885 (2006)
Cavalier-Smith, T. Eukaryotes with no mitochondria. Nature 326, 332–333 (1987)
Cavalier-Smith, T. in Endocytobiology II (eds Schwemmler, W. & Schenk, H.E.A. ) 1027–1034 (De Gruyter, 1983)
van der Giezen, M., Tovar, J. & Clark, C. G. Mitochondria-derived organelles in protists and fungi. Int. Rev. Cytol. 244, 175–225 (2005)
Andersson, S. G. et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396, 133–140 (1998)
Horner, D. S., Hirt, R. P., Kilvington, S., Lloyd, D. & Embley, T. M. Molecular data suggest an early acquisition of the mitochondrion endosymbiont. Proc. R. Soc. Lond. B 263, 1053–1059 (1996)
Lane, N. & Martin, W. The energetics of genome complexity. Nature 467, 929–934 (2010)
Martin, W. & Koonin, E. V. Introns and the origin of nucleus-cytosol compartmentalization. Nature 440, 41–45 (2006)
Lombard, J., Lopez-Garcia, P. & Moreira, D. The early evolution of lipid membranes and the three domains of life. Nature Rev. Microbiol. 10, 507–515 (2012)
Pitcher, A. et al. Core and intact polar glycerol dibiphytanyl glycerol tetraether lipids of ammonia-oxidizing archaea enriched from marine and estuarine sediments. Appl. Environ. Microbiol. 77, 3468–3477 (2011)
van de Vossenberg, J. L., Driessen, A. J. & Konings, W. N. The essence of being extremophilic: the role of the unique archaeal membrane lipids. Extremophiles 2, 163–170 (1998)
Boucher, Y., Kamekura, M. & Doolittle, W. F. Origins and evolution of isoprenoid lipid biosynthesis in archaea. Mol. Microbiol. 52, 515–527 (2004)
Lombard, J., Lopez-Garcia, P. & Moreira, D. An ACP-independent fatty acid synthesis pathway in archaea: implications for the origin of phospholipids. Mol. Biol. Evol. 29, 3261–3265 (2012)
Guldan, H., Matysik, F. M., Bocola, M., Sterner, R. & Babinger, P. Functional assignment of an enzyme that catalyzes the synthesis of an archaea-type ether lipid in bacteria. Angew. Chem. Int. Edn Engl. 50, 8188–8191 (2011)
Tan, H. H., Makino, A., Sudesh, K., Greimel, P. & Kobayashi, T. Spectroscopic evidence for the unusual stereochemical configuration of an endosome-specific lipid. Angew. Chem. Int. Edn Engl. 51, 533–535 (2012)
Shimada, H. & Yamagishi, A. Stability of heterochiral hybrid membrane made of bacterial sn-G3P lipids and archaeal sn-G1P lipids. Biochemistry 50, 4114–4120 (2011)Reports the production of stable heterochiral membranes containing a mixture of bacterial- and archaeal-type lipids, demonstrating the feasibility of natural mixed membranes.
Martin, W. & Muller, M. The hydrogen hypothesis for the first eukaryote. Nature 392, 37–41 (1998)
Nelson-Sathi, S. et al. Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc. Natl Acad. Sci. USA 109, 20537–20542 (2012)
Hampl, V. et al. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc. Natl Acad. Sci. USA 106, 3859–3864 (2009)
Song, S., Liu, L., Edwards, S. V. & Wu, S. Resolving conflict in eutherian mammal phylogeny using phylogenomics and the multispecies coalescent model. Proc. Natl Acad. Sci. USA 109, 14942–14947 (2012)
Lindås, A. C., Karlsson, E. A., Lindgren, M. T., Ettema, T. J. & Bernander, R. A unique cell division machinery in the Archaea. Proc. Natl Acad. Sci. USA 105, 18942–18946 (2008)
Makarova, K. S., Yutin, N., Bell, S. D. & Koonin, E. V. Evolution of diverse cell division and vesicle formation systems in Archaea. Nature Rev. Microbiol. 8, 731–741 (2010)
Blombach, F. et al. Identification of an ortholog of the eukaryotic RNA polymerase III subunit RPC34 in Crenarchaeota and Thaumarchaeota suggests specialization of RNA polymerases for coding and non-coding RNAs in Archaea. Biol. Direct 4, 39 (2009)
Daniels, J. P., Kelly, S., Wickstead, B. & Gull, K. Identification of a crenarchaeal orthologue of Elf1: implications for chromatin and transcription in Archaea. Biol. Direct 4, 24 (2009)
Rivera, M. C. & Lake, J. A. Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science 257, 74–76 (1992)
Acknowledgements
This work was supported by a Marie Curie postdoctoral fellowship to T.A.W. T.M.E. acknowledges support from the European Research Council Advanced Investigator Programme and the Wellcome Trust. We thank J. Archibald for comments on the manuscript.
Author information
Authors and Affiliations
Contributions
T.A.W., P.G.F., C.J.C. and T.M.E. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-2 and additional references. (PDF 185 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Williams, T., Foster, P., Cox, C. et al. An archaeal origin of eukaryotes supports only two primary domains of life. Nature 504, 231–236 (2013). https://doi.org/10.1038/nature12779
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12779
This article is cited by
-
Mysterious Asgard archaea microbes reveal their inner secrets
Nature (2023)
-
Inference and reconstruction of the heimdallarchaeial ancestry of eukaryotes
Nature (2023)
-
Eukaryogenesis and oxygen in Earth history
Nature Ecology & Evolution (2022)
-
Expanding Asgard members in the domain of Archaea sheds new light on the origin of eukaryotes
Science China Life Sciences (2022)
-
The Origin(s) of Cell(s): Pre-Darwinian Evolution from FUCAs to LUCA
Journal of Molecular Evolution (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.