Abstract
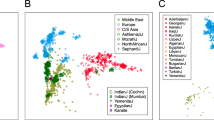
Contemporary Jews comprise an aggregate of ethno-religious communities whose worldwide members identify with each other through various shared religious, historical and cultural traditions1,2. Historical evidence suggests common origins in the Middle East, followed by migrations leading to the establishment of communities of Jews in Europe, Africa and Asia, in what is termed the Jewish Diaspora3,4,5. This complex demographic history imposes special challenges in attempting to address the genetic structure of the Jewish people6. Although many genetic studies have shed light on Jewish origins and on diseases prevalent among Jewish communities, including studies focusing on uniparentally and biparentally inherited markers7,8,9,10,11,12,13,14,15,16, genome-wide patterns of variation across the vast geographic span of Jewish Diaspora communities and their respective neighbours have yet to be addressed. Here we use high-density bead arrays to genotype individuals from 14 Jewish Diaspora communities and compare these patterns of genome-wide diversity with those from 69 Old World non-Jewish populations, of which 25 have not previously been reported. These samples were carefully chosen to provide comprehensive comparisons between Jewish and non-Jewish populations in the Diaspora, as well as with non-Jewish populations from the Middle East and north Africa. Principal component and structure-like analyses identify previously unrecognized genetic substructure within the Middle East. Most Jewish samples form a remarkably tight subcluster that overlies Druze and Cypriot samples but not samples from other Levantine populations or paired Diaspora host populations. In contrast, Ethiopian Jews (Beta Israel) and Indian Jews (Bene Israel and Cochini) cluster with neighbouring autochthonous populations in Ethiopia and western India, respectively, despite a clear paternal link between the Bene Israel and the Levant. These results cast light on the variegated genetic architecture of the Middle East, and trace the origins of most Jewish Diaspora communities to the Levant.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Ben-Sasson, H. H. A History of the Jewish People (Harvard Univ. Press, 1976)
De Lange, N. Atlas of the Jewish World (Phaidon Press, 1984)
Mahler, R. A History of Modern Jewry (Schocken, 1971)
Stillman, N. A. Jews of Arab Lands: A History and Source Book (Jewish Publication Society of America, 1979)
Della Pergola, S. in Papers in Jewish Demography 1997 (eds Della Pergola, S. & Even, J.) 11–33 (The Hebrew University of Jerusalem, 1997)
Cavalli-Sforza, L. L., Menozzi, A. & Piazza, A. in The History and Geography of Human Genes 4 (Princeton Univ. Press, 1994)
Bauchet, M. et al. Measuring European population stratification with microarray genotype data. Am. J. Hum. Genet. 80, 948–956 (2007)
Behar, D. M. et al. Counting the founders: the matrilineal genetic ancestry of the Jewish Diaspora. PLoS ONE 3, e2062 (2008)
Hammer, M. F. et al. Jewish and Middle Eastern non-Jewish populations share a common pool of Y-chromosome biallelic haplotypes. Proc. Natl Acad. Sci. USA 97, 6769–6774 (2000)
Kopelman, N. M. et al. Genomic microsatellites identify shared Jewish ancestry intermediate between Middle Eastern and European populations. BMC Genet. 10, 80 (2009)
Need, A. C., Kasperaviciute, D., Cirulli, E. T. & Goldstein, D. B. A genome-wide genetic signature of Jewish ancestry perfectly separates individuals with and without full Jewish ancestry in a large random sample of European Americans. Genome Biol. 10, R7 (2009)
Olshen, A. B. et al. Analysis of genetic variation in Ashkenazi Jews by high density SNP genotyping. BMC Genet. 9, 14 (2008)
Ostrer, H. A genetic profile of contemporary Jewish populations. Nature Rev. Genet. 2, 891–898 (2001)
Price, A. L. et al. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 4, e236 (2008)
Seldin, M. F. et al. European population substructure: clustering of northern and southern populations. PLoS Genet. 2, e143 (2006)
Tian, C. et al. Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genet. 4, e4 (2008)
Abdulla, M. A. et al. Mapping human genetic diversity in Asia. Science 326, 1541–1545 (2009)
Li, J. Z. et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319, 1100–1104 (2008)
Jakobsson, M. et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature 451, 998–1003 (2008)
Novembre, J. et al. Genes mirror geography within Europe. Nature 456, 98–101 (2008)
Reich, D., Thangaraj, K., Patterson, N., Price, A. L. & Singh, L. Reconstructing Indian population history. Nature 461, 489–494 (2009)
Biswas, S., Scheinfeldt, L. B. & Akey, J. M. Genome-wide insights into the patterns and determinants of fine-scale population structure in humans. Am. J. Hum. Genet. 84, 641–650 (2009)
Tishkoff, S. A. et al. The genetic structure and history of Africans and African Americans. Science 324, 1035–1044 (2009)
Patterson, N., Price, A. L. & Reich, D. Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006)
Hourani, A. A History of the Arab Peoples (Faber & Faber, 1991)
Weiss, K. M. & Long, J. C. Non-Darwinian estimation: my ancestors, my genes’ ancestors. Genome Res. 19, 703–710 (2009)
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009)
Rasmussen, M. et al. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature 463, 757–762 (2010)
Gao, X. & Martin, E. R. Using allele sharing distance for detecting human population stratification. Hum. Hered. 68, 182–191 (2009)
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007)
Acknowledgements
We thank the individuals who provided DNA samples for this study, including the National Laboratory for the Genetics of Israeli Populations; Mari Nelis, Georgi Hudjashov and Viljo Soo for conducting the autosomal genotyping; Lauri Anton for computational help. R.V. and D.M.B. thank the European Commission, Directorate-General for Research for FP7 Ecogene grant 205419. R.V. thanks the European Union, Regional Development Fund through a Centre of Excellence in Genomics grant and the Swedish Collegium for Advanced Studies for support during the initial stage of this study. E.M. and Si.R. thank the Estonian Science Foundation for grants 7858 and 7445, respectively. K.S. thanks the Arthur and Rosalinde Gilbert Foundation fund of the American Technion Society. Sa.R. thanks the European Union for Marie Curie International Reintegration grant CT-2007-208019, and the Israeli Science Foundation for grant 1227/09. IPATIMUP is an Associate Laboratory of the Portuguese Ministry of Science, Technology and Higher Education and is partly supported by Fundação para a Ciência ea Tecnologia, the Portuguese Foundation for Science and Technology.
Author information
Authors and Affiliations
Contributions
D.M.B. and R.V. conceived and designed the study. B.B.T., D.C., D.G., D.M.B., E.K.K., G.C., I.K., L.P., M.F.H., O.B., O.S., T.P. and R.V. provided DNA samples to this study. E.M., J.P. and G.Y. screened and prepared the samples for the autosomal genotyping. D.M.B., E.M., G.C., M.F.H. and Si.R. generated and summarized the database for the uniparental analysis. B.Y., M.M. and Sa.R. designed and applied the modelling methodology and statistical analysis. T.P. provided expert input regarding the relevant historical aspects. B.Y., D.M.B., K.S., M.F.H., M.M., R.V. and Sa.R. wrote the paper. B.Y., D.M.B. and M.M. contributed equally to the paper. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Notes 1-6, References and Supplementary Tables 1-5. (PDF 1143 kb)
Supplementary Figures
This file contains Supplementary Figures 1 and 3-6 and legends for Supplementary Figures 1-6 (see separate file for Supplementary Figure 2) (PDF 6701 kb)
Supplementary Figure 2
This file shows the Principal Component Analysis of the Old World High-Density Array Data. a, Scatter plot of Old World individuals, showing the first two principal components. Here, the first PC (4.2% of variation, vertical axis) captures primarily differences between sub-Saharan Africans and the rest of the Old World. The second PC (3.4% of variation, horizontal axis) differentiates West Eurasians from South and East Asians. Axes of variation were scaled according to eigenvalues. Each letter code (Supplementary Table 1) corresponds to one individual and the colour indicates population origin. b, Scatter plot of Old World individuals, showing PC1 and PC3. c, Scatter plot of Old World individuals, showing PC1 and PC4. Note that eigenvalues for PC3 and PC4 are ~8 times smaller than for PC1 and 2. (PDF 1892 kb)
Rights and permissions
About this article
Cite this article
Behar, D., Yunusbayev, B., Metspalu, M. et al. The genome-wide structure of the Jewish people. Nature 466, 238–242 (2010). https://doi.org/10.1038/nature09103
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09103
This article is cited by
-
Smoking changes adaptive immunity with persistent effects
Nature (2024)
-
Exploring regional aspects of 3D facial variation within European individuals
Scientific Reports (2023)
-
Recurring pathogenic variants in the BRCA2 gene in the Ethiopian Jewish population. Founder mutations?
Familial Cancer (2022)
-
A common founder effect of the splice site variant c.-23 + 1G > A in GJB2 gene causing autosomal recessive deafness 1A (DFNB1A) in Eurasia
Human Genetics (2022)
-
The opposing trends of body mass index and blood pressure during 1977–2020; nationwide registry of 2.8 million male and female adolescents
Cardiovascular Diabetology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.