Abstract
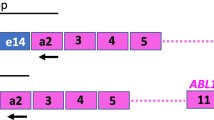
We compared quantitative reverse transcriptase polymerase chain reaction (Q-RT-PCR) to qualitative RT-PCR in determining response to therapy and predicting clinical outcome in 18 retrospectively selected patients with ALL positive for the ALL1-AF4 fusion and with frozen RNA samples collected at diagnosis and during follow-up (96 samples analysed). The ALL1-AF4 junction was detected by qualitative RT-PCR in 18 patients and by Q-RT-PCR in 17 patients (one patient harboured the rare e10-e6 ALL1-AF4 junction, which falls outside of the primer and probe location designed for the Q-RT-PCR). In three of the 12 patients negative to qualitative RT-PCR after induction therapy, a small number of ALL1-AF4 copies was detected by Q-RT-PCR. Thus nine patients were negative and eight positive. Seven of the eight positive patients suffered a relapse, including two of the three patients positive to Q-RT-PCR yet negative to qualitative RT-PCR. Moreover, we found two (5%) discordant results among the 39 follow-up tests of the nine patients who converted to a negative qualitative–quantitative PCR status. The results suggest that qualitative RT-PCR is more appropriate for the routine diagnosis of this genetic alteration. However, Q-RT-PCR is more accurate in assessing the molecular response after induction treatment and could be more useful in clinical decision-making in ALL1-AF4-positive ALL patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Bustin SA . Review. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 2000; 25: 169–193.
Gabert J . Detection of recurrent translocations using real time PCR assessment of the technique for diagnosis and detection of minimal residual disease. Haematologica 1999; 84: 107–109.
Amabile M, Giannini B, Testoni N, Montefusco V, Rosti G, Zardini C et al. Real-time quantification of different types of bcr-abl transcript in chronic myeloid leukemia. Haematologica 2001; 86: 252–259.
Neumann F, Herold C, Hildebrandt B, Kobbe G, Aivado M, Rong A et al. Quantitative real-time reverse-transcription polymerase chain reaction for diagnosis of BCR-ABL positive leukemias and molecular following allogeneic stem cell transplantation. Eur J Haematol 2003; 70: 1–10.
Viehmann S, Teigler-Schlegel A, Bruch J, Langebrake C, Reinhardt D, Harbott J . Monitoring of minimal residual disease (MRD) by real-time quantitative reverse transcription PCR (RQ-RT-PCR) in childhood acute myeloid leukemia with AML1/ETO rearrangement. Leukemia 2003; 17: 1130–1136.
Guerrasio A, Pilatrino C, De Micheli D, Cilloni D, Serra A, Gottardi E et al. Assessment of minimal residual disease (MRD) in CBFbeta/MYH11-positive acute myeloid leukemias by qualitative and quantitative RT-PCR amplification of fusion transcripts. Leukemia 2002; 16: 1176–1181.
Nucifora G, Larson RA, Rowley JD . Persistence of the t(8;21) in patients with acute myeloid leukemia type M2 in long term remission. Blood 1993; 82: 712–715.
Kusec R, Laczika K, Knobl P, Friedl J, Greinix H, Kahals P et al. AML1/ETO fusion mRNA can be detected in remission blood samples of all patients with t(8;21)acute myeloid leukemia after chemotherapy or autologous bone marrow transplantation. Leukemia 1994; 8: 735–739.
Lo Coco F, Diverio D, Pandolfi PP, Biondi A, Rossi V, Avvisati G et al. Molecular evaluation of residual disease as a predictor of relapse in acute promyelocytic leukemia. Lancet 1992; 340: 1437–1438.
Diverio D, Pandolfi PP, Biondi A, Avvisati G, Petti MC, Mandelli F et al. Absence of reverse transcription-polymerase chain reaction detectable residual disease in patients with acute promyelocytic leukemia in long-term remission. Blood 1993; 82: 3556–3559.
Cimino G, Elia L, Rivolta A, Rapanotti MC, Rossi V, Alimena G et al. Clinical relevance of residual disease monitoring by polymerase chain reaction in patients with ALL1-AF4 positive-acute lymphoblastic leukemia. Br J Haematol 1996; 92: 659–664.
Annino L, Vegna ML, Camera A, Specchia G, Visani G, Fioritoni G, et al, for the Gimema group. Treatment of adult acute lymphoblastic leukemia (ALL): long-term follow-up of the GIMEMA ALL 0288 randomized study. Blood 2002; 99: 863–871.
Cimino G, Elia L, Mancini M, Annino L, Anaclerico B, Fazi P, et al, for the GIMEMA group. Clinico-biologic features and treatment outcome of adult pro-B-ALL patients enrolled in the GIMEMA 0496 study: absence of the ALL1/AF4 and of the BCR/ABL fusion genes correlates with a significantly better clinical outcome. Blood 2003; 102: 2014–2020.
Aricò M, Conter V, Valsecchi MG, Rizzari C, Barisone E . Treatment results of high-risk defined by NCI criteria childhood acute lymphoblastic leukemia in AIEOP-ALL studies over two decades. Blood 2000; 96: 464–465, (abstract).
Mitelman F . ISCN (1995). An International System for Human Cytogenetic Nomenclature. Basel: S Karger, 1995.
Chomczynski P, Sacchi N . Single step method of RNA isolation by acid guanidium thiocyanate–phenol–chloroform extraction. Anal Biochem 1987; 162: 156–159.
Biondi A, Rambaldi A, Rossi V, Elia L, Caslini C, Basso G et al. Detection of ALL-1/AF4 fusion transcript by reverse transcription–polymerase chain reaction for diagnosis and monitoring of acute leukemias with the t(4;11) translocation. Blood 1993; 82: 2943–2947.
Cimino G, Elia L, Rapanotti MC, Sprovieri T, Mancini M, Cuneo A et al. A prospective study of residual-disease monitoring of the ALL1/AF4 transcript in patients with t(4;11) acute lymphoblastic leukemia. Blood 2000; 95: 96–101.
Gabert J, Beillard C, van der Velden VHJ, Bi W, Grimwade D, Pallisgaard N et al. Standardization and quality control studies of ‘real-time’ quantitative reverse transcriptase polymerase chain reaction (RQ-PCR) of fusion gene transcripts for residual disease detection in leukemia – a Europe Against Cancer Program. Leukemia 2003; 17: 2318–2357.
Beillard E, Pallisgaard N, van der Velden VH, Bi W, Dee R, van der Schoot E et al. Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using ‘real-time’ quantitative reverse-transcriptase polymerase chain reaction (RQ-PCR) – a Europe Against Cancer program. Leukemia 2003; 17: 1–13.
Miller SA, Dykes DD, Polesky HF . A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16: 1215.
Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G et al. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL1-gene, related to Drosophila trithorax to AF4 gene. Cell 1991; 71: 701–708.
van Dongen JJ, Macintyre EA, Gabert JA, Delabesse E, Rossi V, Saglio G et al. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Leukemia 1999; 13: 1901–1928.
Pallisgaard N, Clausen N, Schroder H, Hokland P . Rapid and sensitive minimal residual disease detection in acute leukemia by quantitative real-time RT-PCR exemplified by t(12;21) TEL/AML1 fusion transcript. Genes Chromosomes Cancer 1999; 26: 355–365.
Bustin SA . Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol 2002; 29: 23–39.
Siraj A K, Ozbek U, Sazawal S, Sirma S, Timson G, Al-Nasser A et al. Preclinical validation of a monochrome real-time multiplex assay for translocations in childhood acute lymphoblastic leukemia. Clin Cancer Res 2002; 8: 3832–3840.
Gallagher RE, Yeap BY, Bi W, Livak KJ, Beaubier N, Rao S et al. Quantitative real-time RT-PCR analysis of PML-RARα mRNA in acute promyelocytic leukemia: assessment of prognostic significance in adult patients from intergroup protocol 0129. Blood 2003; 101: 2521–2528.
Schnittger S, Weisser M, Schoch C, Hiddemann W, Haferlach T, Kern W . New score predicting for prognosis in PML-RARA+,AML1-ETO+, or CBFB-MYH11+ acute myeloid leukemia based on quantification of fusion transcripts. Blood 2003; 102: 2746–2755.
Lavau C, Szilvassy SJ, Slany R, Clery ML . Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J 1997; 16: 4226–4237.
Caslini C, Alarcon AS, Hess JL, Tanaka R, Murti KG, Biondi A . The amino terminus targets the mixed lineage leukemia (MLL) protein to the nucleolus, nuclear matrix and mitotic chromosomal scaffolds. Leukemia 2000; 14: 1898–1908.
Mori H, Colman SM, Xiao Z, Ford AM, Healy LE, Donaldson C et al. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proc Natl Acad Sci USA 2002; 99: 8242–8247.
Bose S, Deininger M, Gora-Tybor J, Goldman JM, Melo JV . The presence of typical and atypical BCR-ABL fusion genes in leukocytes of normal individuals: biologic significance and implications for the assessment of minimal residual disease. Blood 1998; 92: 3362–3367.
Acknowledgements
This work was supported by the Associazione Italiana contro le Leucemie – Sezione di Roma (ROMAIL-ONLUS), Consiglio Nazionale delle ricerche (CNR), Legge 27 dicembre 1997, no. 449/97 and by Ospedale Bambino Gesù-IRCCS Grant 98/02/P/480.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Elia, L., Gottardi, E., Floriddia, G. et al. Retrospective comparison of qualitative and quantitative reverse transcriptase polymerase chain reaction in diagnosing and monitoring the ALL1-AF4 fusion transcript in patients with acute lymphoblastic leukaemia. Leukemia 18, 1824–1830 (2004). https://doi.org/10.1038/sj.leu.2403448
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.leu.2403448