Abstract
In the past decades, the Tai people are increasingly being focused by genetic studies. However, a systematic genetic study of the whole Tai people is still lacking, thus making the population structure as well as the demographic history of this group uninvestigated from genetic perspective. Here we extensively analyzed the variants of hypervariable segments I and II (HVS-I and HVS-II) of mitochondrial DNA (mtDNA) of 719 Tai samples from 19 populations, covering virtually all of the current Tai people’s residences. We observed a general close genetic affinity of the Tai people, reflecting a common origin of this group. Taken into account the phylogeographic analyses of their shared components, including haplogroups F1a, M7b and B5a, our study supported a southern Yunnan origin of the Tai people, consistent with the historical records. In line with their diverse cultures and languages, substantial genetic divergences can be observed among different Tai populations that could be attributable to assimilation of maternal components from neighboring populations. Our study further implied the advent of rice agriculture in Mainland Southeast Asia at ∼5 kya (kilo years ago) had greatly promoted the population expansion of the Tai people.
Similar content being viewed by others
Introduction
The Tai people, also called Tai-Lao or Tai-Shan group, refer to the populations that descended from a common ancestor speaking the Proto-Tai languages.1 With a large population size (∼60 million), this group is now living in Indo-China peninsula, including Dai people in China, Thai and Phuthai people in Thailand, Lao people in Laos, Shan people in Myanmar and Ahom people in northeast India (NEI). All of the contemporary Tai populations can be assigned to the southwestern branch of the Tai language (http://www.ethnologue.com/)2 that belongs to the Tai-Kadai language family. Considering their language similarities with the other Tai-Kadai populations, it is now widely accepted that the Tai people could trace their origin back to the ancient Bai-Yue tribe inhabiting in south and southeast China at ∼2000–3000 years ago.3, 4 At the end of Tang and Song Dynasty, some Tai people migrated southward into Mainland Southeast Asia (MSEA), probably compelled by political and cultural compression from northern China.3 During their formation history, the Tai people assimilated cultures from the other civilizations in MSEA and NEI, and evolved into different Tai populations with diverse cultural and linguistic characters. For example, Tais from different regions could be assigned to different language subbranches, including Tai Lü, Tai Nüa, Tai Hongjin, Tai Ya, Tai Dam, Tai Dón and Ahom languages.2 Meanwhile, the cultures of these Tai populations, including the custom and the dressing characteristics, are quite different, even for Dai populations residing in Xishuangbanna and Dehong.4 Therefore, it would be an interesting issue to explore whether the current Tai people maintained any common genetic basis, or whether their genetic compositions had been affected by the neighboring populations during their cultural and linguistic differentiation.
In the past two decades, substantial efforts have been carried out to unravel the ethnohistory of the Tai people from the perspective of genetics. However, these genetic studies focused either on the genetic composition of a certain Tai population, such as Dais from Xishuangbanna,5, 6, 7 Thais from Thailand,8, 9, 10 Shans from Myanmar,11 Lao people from Cambodia12 and Ahom people from NEI,13 or the genetic relationship of the Tais with other linguistic populations5 or the effect of mitochondrial DNA (mtDNA) haplogroup on drug addiction and HIV-1 infection of Dai samples.14 Some Tai populations, including the normal Dais from Dehong, Lincang, Lancang, Yuanjiang and Honghe in Yunnan, China, remain uninvestigated.
To systematically study the genetic landscape of the whole Tai people and reconstruct their ethnohistory, mtDNA variations of 19 Tai populations that covered virtually all the current distribution regions of the Tai people are collected and investigated here, with particular attempt to address the question about how the current Tai populations originated and evolved during their long formation history.
Materials and methods
Samples
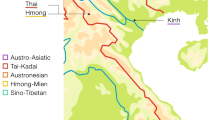
A total of 264 Dai individuals from 7 populations residing in Yunnan are newly collected in the present study (Figure 1). These populations are from Xishuangbanna Dai Autonomous Prefecture (Dai_6), Dehong Dai and Jingpo Autonomous Prefecture (Dai_3 and Dai_4), Lancang (Dai_5), Lincang (Dai_9), Yuanjiang Hani, Yi and Dai Autonomous County (Dai_7) and Honghe Hani and Yi Autonomous Prefecture in Jianshui (Dai_8). The mtDNA data of Tai people are also collected from the literature, including Dai people from Xishuangbanna,5, 6, 7 Shan people from Myanmar,11 Thai people and Phuthai people from Thailand,8, 9, 10 Lao people from Cambodia12 and Ahom people from NEI.13 In total, 719 Tai samples from 19 populations are studied in this study, covering virtually all the residing areas of the Tai people, including southwestern China, MSEA and NEI (Figure 1 and Table 1).
To unravel the genetic relationship of the Tai populations with their neighboring populations, previously published mtDNA data, including 3499 Tibeto-Burmans (TB), 2172 Austro-Asiatics (AA), 1030 Tai-Kadais (TK) and 633 Hmong-Miens (HM), are also collected for comparison (Figure 1 and Supplementary Table S1). Considering the genetic differentiation of the AA populations observed in previous studies,15, 16 the AA populations are further divided into Mon-Khmer and Munda groups in our analyses.
Sequencing and haplogroup allocation
Procedures for the extraction of genomic DNA, PCR amplification and DNA sequencing of mtDNA hypervariable segments (HVS) have already been well described elsewhere.17, 18, 19 The 264 new mtDNA sequences reported in this study have been deposited in GenBank under accession numbers (KU862353–KU862616). All samples were first assigned into respective haplogroups on the basis of control region and partial coding region information according to the reconstructed mtDNA tree (http://phylotree.org/). For the rest of unassigned mtDNAs, motif search20 in large-scale reported data set harboring >55 000 mtDNA HVS data21, 22 was carried out, with the special attention to identify all the possible existed basal lineages in the samples with minimum efforts. Based on the phylogenetic tree, certain coding region sites were selected for typing by either restriction fragment length polymorphism or DNA sequencing in order to confirm the haplogroup status inferred from the control region motifs. The haplogrouping of the mtDNA sequences were further checked by using MitoTool.23
Data analyses
The principal component analysis (PCA) was performed based on haplogroup frequency matrix (Supplementary Table S2) by using the Statistical Package for the Social Sciences (SPSS, Chicago, IL, USA) software (version 16.0), whereas the contribution of each haplogroup to the clustering pattern was calculated as the factor scores for the first and the second principal components (PCs) with regression (REGR) method in SPSS software. Contour maps of spatial frequencies of haplogroup were constructed using Kriging linear model of Surfer 8.0 (Golden Software Inc., Golden, CO, USA). Reduced median network for each haplogroup was constructed based on HVS-I data by the program NETWORK 5.0 (www.fluxus-engineering.com/sharenet.htm).24 Coalescent ages of haplogroups were calculated by rho (ρ) statistic method.25, 26 Mutation rates of transitions between 16 090 and 16 365, and substitutions between 16 051 and 16 400,27 were adopted for age estimations. FST between populations were calculated by using the package Arlequin 3.11.28 Bayesian skyline plot (BSP) for effective population size (Nef) through time was reconstructed based on the HVS-I data (16 051–16 400) using BEAST V 1.5.2.29 In the BSP analysis, the best-fit model was selected with Model test (version 3.7),30 a strict molecular clock with the fixed rate as 1.691e−8 substitutions per site per year31 was chosen. Each Markov chain Monte Carlo32, 33 was run for 40 000 000 generations and sampled every 4000 generations, with the first 40 000 generations discarded as burn-in. The results were visualized with Tracer v1.5 (http://tree.bio.ed.ac.uk/software/tracer/).
Results
Haplogroup profile of the Tai samples
Among the 264 Dai individuals, the majority (241/264, 91.29%) could be classified into eastern Eurasian haplogroups, such as A, B, D, G, M7, M8, M9, M10, M12, N9 and R9 (Supplementary Table S3 and Supplementary Figure S1). Paleolithic lineages that are mainly found in southern East Asia and Southeast Asia,34 including M20, M24, M71, M74, N21 and R11, are rare in these Dai samples (13/264, 4.92%). Haplogroups that are prevalent in South Asia, for example, M5, M33 and M60, accounted for only 1.89% of the samples. The high proportion of east Eurasian components remains stable (610/719, 84.84%) after taking into account all the 19 Tai populations (Supplementary Figure S1), similar with the maternal genetic structure of other Tai-Kadai groups.5, 6, 7, 8, 9, 10 Considering the absence or low frequencies of these South Asian lineages and southern East Asian Paleolithic components in the majority of the studied Tai populations, it is probable that the presence of these lineages in Tai people would have resulted from recent gene flow.
The genetic affinity among Tai populations
The clustering pattern of the Tai populations and other groups such as TB, HM, AA and other TK is displayed in the PC plot (Figure 2a). A general division between TB and southern East Asian groups (including Mon-Khmer, HM and TK populations) could be observed based on PC1, in line with the previous observations.22, 35 Haplogroups F1a, B4, B5a and M7b contributed most to the southern East Asian cluster (Figure 2b). On the contrary, haplogroups M9, A and D contributed most to the TB groups, in which Munda and Ahom populations from NEI concentrated. This genetic differentiation could be further supported by the greater FST values between Tais and TB (Supplementary Figure S2d) as well as Munda (Supplementary Figure S2f) groups, and the analysis of molecular variance result (P<0.05; data not shown). With the exception of Ahom from NEI and Shan from Myanmar that mixed with or is slightly shifted towards the TB group (Supplementary Figure S2d), the majority of the Tai populations clustered together with each other and furthermore fell within the southern East Asian groups. The close clustering between different Tais could also be supported by their close pairwise genetic distances (Supplementary Figure S2a), thus reflecting an ethnolinguistic affinity.
PCA of populations analyzed in the present study. (a) PC map of the 152 populations based on haplogroup frequencies. The 19 Tai populations are labeled. (b) Plot of the haplogroup contribution of the first and second PCs. The contribution of each haplogroup was calculated as the factor scores for PC1 and PC2 with REGR method in SPSS. A full color version of this figure is available at the Journal of Human Genetics journal online.
The shared genetic components among Tai people
The close genetic affinity of different Tai populations, as indicated by their clustering pattern in the PC map (Figure 2a) and small FST values (Supplementary Figure S2a), suggest that the common genetic basis of this group is retained in the contemporary populations. Among the haplogroups that contributed most to the Tai people (Figure 2b), haplogroups F1a, M7b and B5a accounted for 33.38% (240/719) of our Tai samples and distributed widely in almost all of these populations. These shared haplogroups would have witnessed the origin and migration of the Tai people and thus will shade light on the ethnohistory of this group.
Age estimations revealed that the dates of these haplogroups (and their major nodes) could be divided into three periods, late Paleolithic period (23–12 kya (kilo years ago)), early Neolithic period (12–7 kya) and Bronze or Ages (2–3 kya), thus implying a genetic stratigraphy of the Tai people (Table 2). In detail, F1a, F1a1 and M7b that could be traced back to ∼23–19 kya might represent genetic relics of the Last Glacial Maximum (LGM; ∼26.5–19 kya).36 Other late Paleolithic nodes, such as F1a*, F1a1a* and B5a* with coalescent ages ranging from 17.53 to 11.05 kya, indicated a population expansion during the post-glacial era. Haplogroups F1a1*, M7b1a1*, M7b1a1a* and M7b1a1-16192*, which coalesced ∼7–12 kya and had star-like structures, would reflect demographic expansion during the Holocene period. Differently, the ages of other two nodes, M7b1a1* and M7b1a1-16192*, are much younger (∼2–3 kya), possibly reflecting the prosperity of the Tai populations during Bronze and Iron Ages.
Contour maps of these haplogroups based on large-scale data set that covered almost the whole Asia revealed southern East Asian (especially southern China) distributions of their ancestor nodes (including F1a*, F1a1*, B5a* and M7b*; Figure 3), thus implying their potential origins in this area. Moreover, the emergences of the terminal nodes (for example, F1a1a, B5a1d and M7b-16189-16192; Figure 3) in the southern regions, especially southern MSEA, indicated their subsequent southward migrations into MSEA. To further explore the origin and expansion history of the Tai people, we reconstructed the networks of these haplogroups based on HVS-I data of Tai samples (Figure 4). Interestingly, the root types of most of these haplogroups, especially the Late Neolithic lineages within the timescale of the origin of the Tai people (for example, F1a1*, M7b1a1a, M7b1a1b, M7b1a-16192 and M7b1a-16189-16192; Figure 4), are mainly occupied by individuals from Yunnan (including Dehong, Lincang, Xishuangbanna, Lancang, Jianshui and Yuanjian). On the contrary, the individuals from NEI, Myanmar, Cambodia and Thailand mainly located sporadically in the terminal branches (Figure 4), reflecting recent immigrations into these areas. Although root types of some haplogroups, including F1a1*, M7b1a1a, M7b1a-16192 and B5a1d, could also be observed in Thailand, Cambodia and even NEI (Figure 4), their fewer sub-branches in these regions made it probable that they immigrated into these places by recent gene flow. These results jointly implied that the ancestors of the Tai people originated in the northern region of their current residences (especially southern Yunnan) after deriving from their ancestors in southern China, and then dispersed southward into MSEA and westward into NEI.
Median networks of haplogroups F1a (a), M7b (b) and B5a (c) based on HVS-I data. Nucleotide position numbers are consistent with the revised Cambridge Reference Sequence (rCRS47). Suffixes A and T refer to transversions; ‘‘@’’ means a reverse mutation. The geographic origin of samples was shown by different colors. The C stretch length polymorphism in regions 303–315, AC indels at 515–522, 16182C, 16183C, 16193.1C(C) and 16519 were disregarded for the network reconstruction. A full color version of this figure is available at the Journal of Human Genetics journal online.
Genetic divergences in Tai populations
Although having a common origin, substantial genetic divergence could also be observed among the Tai people (Figure 2). This is consistent with the different proportions of eastern Eurasian components in the diverse Tai populations (ranging from 30.56 to 100%; Supplementary Figure S1). To dissect the genetic differentiation of the Tai people, we further conducted PCA on TK (including Tai populations), AA and HM populations that clustered together in Figure 2a. Three clusters can be observed in the PC map of these populations (Figure 5a). Consistent with their close ethnolinguistic affinity as observed in Figure 2, most of the Tais, including Shan, Phuthai, Xishuangbanna Dais (Dai_1, Dai_2, Dai_6 and Dai_10), eastern Yunnan Dais (Dai_7 and Dai_8) and Lancang Dai (Dai_5), fell within cluster 1 and genetically related to the other TK and HM populations, in line with the smaller FST values between Tais and these groups (Supplementary Figures S2b and c). Differently, Thais (Thai1-5) from Thailand, Lao from Cambodia as well as Dais from western Yunnan (Dai_3, Dai_4 and Dai_9) are slightly shifted toward the majority of the Mon-Khmers from western Yunnan and MSEA in cluster 2, reflecting genetic exchange between these Tai populations and their surrounding AA groups. This pattern can be further supported by the lower FST between these Tai populations and the Mon-Khmer groups (Supplementary Figure S2e). Similar with Figure 2a, Ahom from NEI positioned far away from the other Tais and fell within cluster 3 that consists mainly of Munda populations from NEI. Taken together with the fact that haplogroup M9 contributed most to this cluster (Figure 5b), and the lower FST between Ahom and both TB (Supplementary Figure S2d) and Munda (Supplementary Figure S2f) populations, it is probable that the Ahom people had assimilated components from their neighboring TB populations during their immigration into this area.
PCA of populations without Tibeto-Burmans. (a) PC map of AA, TK and HM populations based on haplogroup frequencies. (b) Plot of the haplogroup contribution of the first and second PCs. The contribution of each haplogroup was calculated as the factor scores for PC1 and PC2 with REGR method in SPSS. A full color version of this figure is available at the Journal of Human Genetics journal online.
Population dynamics of the Tai people
To further explore the population dynamics of the Tai people, BSP for Nef through time was reconstructed based on the HVS-I data of all the Tai samples. The result suggested a flat growth of the Tai people before 5 kya (Figure 6). After 5 kya, the population size started to increase rapidly and lasted to the present (Figure 6), consistent with the observations in previous study,37, 38 thus indicating a rapid expansion of the Tai people during the late Neolithic period or Iron and Bronze Ages.
BSP of female Nef through time based on HVS-I sequences. The bold black line represents the median population size estimate from Bayesian posterior distribution. The gray lines delimit the 95% highest posterior density (HPD) boundaries, accounting for uncertainty in the reconstructed phylogeny and substitution model parameters. Nef is plotted on a logscale and assumes a female generation time of 20 years.
Discussion
By extensively dissecting the maternal genetic structure of the current Tai people, here we disclosed a close genetic affinity among different Tai populations, reflecting a common origin of this group. Based on phylogeographic analyses on their shared haplogroups, including haplogroups F1a, B5a and M7b, our results indicated a southern China origin of the common ancestors of the Proto-Tais, consistent with their hypothesized Bai-Yue origin.3 Further studies indicated that after evolving from the ancient Bai-Yue tribe, the ancestors of the Tais first occupied southern Yunnan, and then migrated southward into MSEA and NEI. This is in good agreement with historical studies that the Tai language speakers might originate at the northern area of their current locations, including the area ranging from Guangxi, Guizhou, southern Yunnan to northern MSEA.1, 39 The recent immigration of these haplogroups into NEI and southern MSEA (especially Myanmar and Cambodia) fits well with the historical records suggesting that the Tais conquered NEI, Myanmar and Cambodia during the eleventh to the late thirteenth century.40, 41
During their migration, the Tai people assimilated languages and cultures from their neighboring populations, thus evolving into diverse Tai groups. Our study further indicated that this culture differentiation also involved genetic exchanges with their neighboring populations. For example, for the Ahom people from NEI that exhibited as an outlier in the PCA, it is recorded that besides Ahom language, this group also speaks the Assam language that belongs to the Indo-Aryan language family. During their invasion of upper Assam, the Ahom people also subjugated some local TB populations.40 Consequently, the culture of the modern Ahom people converge both the original Tai culture and the Tibeto-Burman culture. It is therefore not surprising that the Ahom people harbored high proportion of Tibeto-Burman prevalent haplogroups (for example, M9) and South Asian lineages (for example, M3, M5, M6, M30, M33 and M49; Supplementary Table S3). Similarly, affiliations between the Shan people and the Myanmar Tibeto-Burmans could be explained by the fact that the Shan people are now speaking both Shan language and Burmese language. In addition, we also noticed that the Dais from western Yunnan, the Lao people from Cambodia and Thais from Thailand are slightly shifted toward their neighboring AA groups, indicating genetic exchanges between these linguistically different populations. This is in line with the linguistic studies suggesting that the Tai populations had assimilated both rice farming technologies and terms from their surrounding AA populations during their formation.42
Interestingly, the advent of rice agriculture probably promoted the population expansion of the Tai people at ∼5 kya. This notion gets further support from the advent of rice agriculture into the Tais’ current locations at <5 kya, as indicated by archaeological records. For example, the fossils of rice grains found in Dadunzi site in Yuanmou, Yunnan, could be dated to ∼4000 BP.43 In addition, the introduction of rice into MSEA is even later,44 as suggested by grain casts in Ban Chiang in northeast Thailand (2000 BCE),45 as well as rice splikelets and chaff found in Khoh Phanom Di site in southern Thailand (1500–2000 BCE).46 Furthermore, given that this expansion time overlapped with the Bronze and Iron Ages in MSEA (1500 BC–500 AD), it is also probable that this rapid expansion could have facilitated the prosperity of the ancient kingdoms, such as the ‘Yue-Shang’ and ‘Da-Guang’ kingdoms established by the Tai people.1
In summary, by systematically studying the genetic landscape of the Tai people, our study revealed a common origin of this group at the northern area of their current locations (especially southern Yunnan). After that, the Tai people assimilated both culture and gene flow from their neighboring populations, thus involving into different Tai groups with diverse cultures and genetic divergences. Considering the wide coverage of our Tai samples, our study is essential in understanding the ethnohistory of the Tai people, as well as the peopling of MSEA from the maternal genetic perspective.
Accession codes
References
He, P. Zhongnanbandao Minzu de Yuanyuan yu Liubian, (The Ethnic Publishing House, Beijing, China, 2006).
Lewis, M. P., Simons, Gary F. & Fennig, Charles D. (eds) Ethnologue: Languages of the World 18th edn (SIL International, Dallas, TX, USA, 2015).
You, Z. History of Yunnan nationalities, (Yunnan University Press, 1994).
Tawakule, W. T. (ed) China's Ethnic Groups, (The Ethnic Publishing House, Beijing, China, 2004).
Yao, Y. G., Nie, L., Harpending, H., Fu, Y. X., Yuan, Z. G. & Zhang, Y. P. Genetic relationship of Chinese ethnic populations revealed by mtDNA sequence diversity. Am. J. Phys. Anthropol. 118, 63–76 (2002).
Qian, Y. P., Chu, Z. T., Dai, Q., Wei, C. D., Chu, J. Y., Tajima, A. et al. Mitochondrial DNA polymorphisms in Yunnan nationalities in China. J. Hum. Genet. 46, 211–220 (2001).
Li, H., Cai, X. Y., Winograd-Cort, E. R., Wen, B., Cheng, X., Qin, Z. D. et al. Mitochondrial DNA diversity and population differentiation in southern East Asia. Am. J. Phys. Anthropol. 134, 481–488 (2007).
Fucharoen, G., Fucharoen, S. & Horai, S. Mitochondrial DNA polymorphisms in Thailand. J. Hum. Genet. 46, 115–125 (2001).
Lertrit, P., Poolsuwan, S., Thosarat, R., Sanpachudayan, T., Boonyarit, H., Chinpaisal, C. et al. Genetic history of Southeast Asian populations as revealed by ancient and modern human mitochondrial DNA analysis. Am. J. Phys. Anthropol. 137, 425–440 (2008).
Jin, H. J., Tyler-Smith, C. & Kim, W. The peopling of Korea revealed by analyses of mitochondrial DNA and Y-chromosomal markers. PLoS ONE 4, e4210 (2009).
Summerer, M., Horst, J., Erhart, G., Weißensteiner, H., Schonherr, S., Pacher, D. et al. Large-scale mitochondrial DNA analysis in Southeast Asia reveals evolutionary effects of cultural isolation in the multi-ethnic population of Myanmar. BMC Evol. Biol. 14, 17 (2014).
Zhang, X. M., Qi, X. B., Yang, Z. H., Serey, B., Sovannary, T., Bunnath, L. et al. Analysis of mitochondrial genome diversity identifies new and ancient maternal lineages in Cambodian aborigines. Nat. Commun 4, 2599 (2013).
Rej, P. H. Measuring Mitochondrial DNA Diversity and Demographic Patterns of Tribal and Caste Populations from the Northeast Indian State of Assam 43–56 (University of Cincinnati, OH, USA, 2013).
Zhang, A. M., Hu, Q. X., Liu, F. L., Bi, R., Yang, B. Q., Zhang, W. et al. Mitochondrial DNA haplogroup a decreases the risk of drug addiction but conversely increases the risk of HIV-1 infection in Chinese addicts. Mol. Neurobiol. (e-pub ahead of print 11 July 2015; doi:10.1007/s12035-015-9323-y).
Chaubey, G., Metspalu, M., Choi, Y., Magi, R., Romero, I. G., Soares, P. et al. Population genetic structure in Indian austroasiatic speakers: the role of landscape barriers and sex-specific admixture. Mol. Biol. Evol. 28, 1013–1024 (2011).
Kumar, V., Reddy, A. N., Babu, J. P., Rao, T. N., Langstieh, B. T., Thangaraj, K. et al. Y-chromosome evidence suggests a common paternal heritage of Austro-Asiatic populations. BMC Evol. Biol. 7, 47 (2007).
Yao, Y. G., Kong, Q. P., Bandelt, H. J., Kivisild, T. & Zhang, Y. P. Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am. J. Hum. Genet. 70, 635–651 (2002).
Kong, Q. P., Yao, Y. G., Liu, M., Shen, S. P., Chen, C., Zhu, C. L. et al. Mitochondrial DNA sequence polymorphisms of five ethnic populations from northern China. Hum. Genet. 113, 391–405 (2003).
Yao, Y. G., Kong, Q. P., Wang, C. Y., Zhu, C. L. & Zhang, Y. P. Different matrilineal contributions to genetic structure of ethnic groups in the silk road region in China. Mol. Biol. Evol. 21, 2265–2280 (2004).
Kong, Q. P., Bandelt, H. J., Sun, C., Yao, Y. G., Salas, A. & Achilli, A. Updating the east asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations. Hum. Mol. Genet 15, 2076–2086 (2006).
Tian, J. Y., Wang, H. W., Li, Y. C., Zhang, W., Yao, Y. G., van Straten, J. et al. A genetic contribution from the Far East into Ashkenazi Jews via the ancient Silk Road. Sci. Rep. 5, 8377 (2015).
Li, Y. C., Wang, H. W., Tian, J. Y., Liu, L. N., Yang, L. Q., Zhu, C. L. et al. Ancient inland human dispersals from Myanmar into interior East Asia since the late Pleistocene. Sci. Rep. 5, 9473 (2015).
Fan, L. & Yao, Y. G. An update to MitoTool: using a new scoring system for faster mtDNA haplogroup determination. Mitochondrion 13, 360–363 (2013).
Bandelt, H. J., Forster, P. & Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 (1999).
Saillard, J., Forster, P., Lynnerup, N., Bandelt, H. J. & Nørby, S. mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am. J. Hum. Genet. 67, 718 (2000).
Forster, P., Harding, R., Torroni, A. & Bandelt, H. J. Origin and evolution of Native American mtDNA variation: a reappraisal. Am. J. Hum. Genet. 59, 935 (1996).
Soares, P., Ermini, L., Thomson, N., Mormina, M., Rito, T., Röhl, A. et al. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am. J. Hum. Genet. 84, 740–759 (2009).
Excoffier, L., Laval, G. & Schneider, S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1, 47 (2005).
Drummond, A. & Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (2007).
Posada, D. & Buckley, T. R. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 53, 793–808 (2004).
Ho, S. Y. W. & Endicott, P. The crucial role of calibration in molecular date estimates for the peopling of the Americas. Am. J. Hum. Genet. 83, 142 (2008).
Metropolis, N., Rosenbluth, A. W., Rosenbluth, M. N., Teller, A. H. & Teller, E. Equation of state calculations by fast computing machines. J. Chem. Phys. 21, 1087–1092 (1953).
Hastings, W. K. Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57, 97–109 (1970).
Kong, Q. P., Sun, C., Wang, H. W., Zhao, M., Wang, W. Z., Zhong, L. et al. Large-scale mtDNA screening reveals a surprising matrilineal complexity in east asia and its implications to the peopling of the region. Mol. Biol. Evol. 28, 513–522 (2011).
Wen, B., Xie, X., Gao, S., Li, H., Shi, H., Song, X. et al. Analyses of genetic structure of Tibeto-Burman populations reveals sex-biased admixture in southern Tibeto-Burmans. Am. J. Hum. Genet. 74, 856–865 (2004).
Clark, P. U., Dyke, A. S., Shakun, J. D., Carlson, A. E., Clark, J., Wohlfarth, B. et al. The last glacial maximum. Science 325, 710 (2009).
Gignoux, C. R., Henn, B. M. & Mountain, J. L. Rapid, global demographic expansions after the origins of agriculture. Proc. Natl Acad. Sci. USA 108, 6044–6049 (2011).
Zheng, H. X., Yan, S., Qin, Z. D., Wang, Y., Tan, J. Z., Li, H. et al. Major population expansion of East Asians began before neolithic time: evidence of mtDNA genomes. PLoS ONE 6, e25835 (2011).
Luo, W., Hartmann, J., Li, J. & Sysamouth, V. GIS mapping and analysis of Tai linguistic and settlement patterns in southern China. GIS 6, 129–136 (2000).
Gait, E. A. A History of Assam, (Thacker, Spink & Company, Harvard University, MA, USA, 1906).
Lieberman, V. B. Strange Parallels: Integration on the Mainland Southeast Asia in global context. (Cambridge University Press: New York, NY, USA, 2003).
Blench, R. in The Peopling of East Asia: Putting Together Archaeology, Linguistics, and Genetics (eds Sagart, L., Blench, R. & Sanchez-Mazas, A.) 40–41 (Routledge, New York, NY, USA, 2005).
Zhimin, A. Origin of chinese rice cultivation and its spread east. Cultural Relics 2, 63–70 (1999).
Weber, S., Lehman, H., Barela, T., Hawks, S. & Harriman, D. Rice or millets: early farming strategies in prehistoric central Thailand. Archaeol. Anthrop. Sci. 2, 79–88 (2010).
Kealhofer, L. Changing perceptions of risk: the development of agro-ecosystems in Southeast Asia. Am. Anthropol. 104, 178–194 (2002).
Thompson, G. B. The Excavation of Khok Phanom Di, a Prehistoric Site in Central Thailand. Volume IV: Subsistence and Environment: The Botanical Evidence (The Biological Remains, Part II), ((Reports of Research Committee of the Society of Antiquaries, London, 1996).
Andrews, R. M., Kubacka, I., Chinnery, P. F., Lightowlers, R. N., Turnbull, D. M. & Howell, N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 23, 147 (1999).
Acknowledgements
We thank the volunteers for participating in this project. This work was supported by grants from the National Natural Science Foundation of China (31322029) to Q-PK, Kunming Institute of Zoology, Chinese Academy of Sciences (GREKF13-09) to WH and the West Light Foundation of The Chinese Academy of Sciences to Y-CL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Li, YC., Huang, W., Tian, JY. et al. Exploring the maternal history of the Tai people. J Hum Genet 61, 721–729 (2016). https://doi.org/10.1038/jhg.2016.36
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2016.36