Abstract
Little information is available regarding the molecular pathogenesis of Parkinson’s disease (PD) among the Bengalee population in West Bengal, India. This study was undertaken to determine the contribution of Parkin variants in well-defined ethnically identical Bengalee population of India and further to describe the clinical spectrum associated with these mutations. A total of 150 unrelated PD patients and 150 controls were recruited for the study. The entire cohort was screened for mutations in all the 12 exons of the gene along with flanking splice junctions by polymerase chain reaction and DNA sequencing. Eleven nucleotide variants including two novel changes were detected. Cerebrospinal fluid (CSF) parkin protein expression of the novel mutation, Val186Ile (found in heterozygous condition in one patient only) was almost 2.7 folds lower than the controls and other PD patients. Molecular characterization of polymorphisms Ser167Asn and Val380Leu depicted that homozygous Ser167 and Val380 are significantly associated with the disease. We did not find any linkage disequilibrium among the SNPs, the low r2 for every pair of single-nucleotide polymorphisms (SNPs) indicated that these SNPs cannot be tagged by each other. Another novel intronic change, IVS8+48C>T was present in almost equally in PD patients and controls. Among the ethnically defined Bengalee population of West Bengal, occurrence of Parkin mutation is 4% (6/150) of the PD patient pool supported with decreased folds of expression of CSF PARKIN protein. Parkin polymorphisms, Ser167 and Val380 are risk factors for the progression of the disease, and their frequency is greatly influenced by ethnic origin.
Similar content being viewed by others
Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative disorder characterized by the symptoms of bradykinesia, tremor, postural instability and muscular rigidity. The past few years have seen great progress in the molecular genetics of PD leading to the identification of different candidate genes (autosomal recessive and dominant). The cause of the degeneration of the dopaminergic nigrostriatal pathway projection in PD is largely unknown although it is assumed to be an effect of complex interaction between genetic and environmental factors. Various biological pathways or processes involved include mitochondrial dysfunction, oxidative damage, abnormal protein accumulation and protein phosphorylation, which may all have a role in dopaminergic neuronal function and survival.1 Mutations in the Parkin gene, an E3 protein-ubiquitin ligase, involved in the ubiquitin–proteasome pathway which results in a loss of function leading to an autosomal form of parkinsonism, are the most common cause of early onset PD in many countries and account for 10–25% of cases. To date, there are more than 366 reported mutations for Parkin gene2 which has been studied in large ethnically mixed patient populations by several groups. However, the reported frequency and spectrum of mutations rate varied by PD sample tested and detection methods used.
Although only three studies previously from various parts of India with hospital-based data have been reported with Parkin as a candidate gene,3, 4, 5 none of them stressed on a particular ethnic population from a specific region of India. Despite such extensive studies, little information is available regarding the expressivity of Parkin protein and molecular pathogenesis of PD among the Bengalee population in West Bengal, India. This study was undertaken to examine the alternate patterns of protein level from cerebrospinal fluid (CSF) and contribution of Parkin variants in well-defined ethnically identical Bengalee population of India, and further to describe the clinical spectrum associated with these mutations.
Subjects and methods
Collection of patient samples
This is a hospital-based study whose study participants belonged to various age and socioeconomic groups. The base population recruited for this study was the outpatients of the Movement Disorder Neuromedicine Clinic visiting the National Neuroscience Centre (NNC) and Nil Ratan Sircar Medical College and Hospital (NRS), Kolkata, India from 1 March 2008 to 28 November 2012. After an initial screening procedure, clinical data and detail family history of each one of the 150 PD patients were collected with the help of collaborating clinicians after physical and neurological examination by two independent consultant neurologists. Hospitals like NRS and NNC are the main referral centers for cases related to movement disorders and are situated in the heart of Kolkata city, West Bengal. We have meticulously selected only those patients whose ancestors were residents of West Bengal, India for the past three generations. The demographic data (name, age, address, place of inhabitation, occupation, family pedigree, educational status, monthly income, occupation/professional status of subject during his/her life time, residence area (rural/urban and so on) were collected by health professionals and epidemiologists. Participants were given an open-ended semi-structured interviewer–administrative questionnaire that collected information on disease history, from disease onset to the baseline evaluation from patients and controls in a face-to-face interview, and a neurologist examined each patient and control face-to-face clinically after obtaining written informed consent. Each interview and evaluation took about 60 to 90 min. All the 300 participants (150 patients with PD and 150 healthy controls) actively took part in this study, and their good response enabled a 100% participation rate. Both participants and the accompanying family members/caregivers were allowed to answer. Before the questionnaire was formally administered, a panel consisting of neurologists, epidemiologists and a biostatistician examined content validity. Patients were classified, based on their history and baseline evaluation, as having an akinetic-dominant, tremor-dominant or mixed tremor/akinetic clinical subtype and symmetrical or asymmetrical symptom onset and progression. In addition, information regarding drug therapy, response to levodopa and demographic variables was collected, including whether patients had previously experienced hallucinations while taking anti-PD medication.
Case definition
In order to achieve high diagnostic specificity as well as high sensitivity, patients with PD were diagnosed according to the criteria of the UK Parkinson’s Disease Society Brain Bank Research, and all the cases had to meet the following criteria of clinical classification of definite, probable, and possible PD at the time of diagnosis and throughout the study period:
-
1
The presence of at least three of the following signs: resting tremor, cogwheel rigidity, bradykinesia and postural reflex impairment, at least one of which must be either rest tremor or bradykinesia. The disease has a unilateral onset and asymmetrical development, and the response to a dopaminergic agent should be good to excellent.
-
2
No suggestion of secondary parkinsonism due to drugs (such as dopamine-blocking or dopamine-depleting agents), trauma, brain tumor or treatment within the last 12 months.
-
3
No atypical features such as prominent oculomotor palsy, cerebellar signs, vocal cord paresis, severe orthostatic hypotension, pyramidal signs, amyotrophy or limb apraxia.
The Unified Parkinson’s Disease Scale,6 Hoehn and Yahr (H&Y)7 staging, activities of daily living, and Mini Mental State Examination8 scale were performed for each patient to estimate the motor symptoms and magnitude of the disease severity.
None of the controls had any diagnosable neurological disorders, cognitive impairment or neuropsychiatric disability in their family history with similar educational levels. One hundred and fifty non-PD control groups consisted of patients’ spouses and other healthy community-based, age–sex-matched volunteers residing in the same ethnic background as the PD patients. The experiments were conducted in accordance with the Declaration of Helsinki. Ethical approval of the research project using human subject was issued from the Institutional Ethical Committee of collaborating hospital and institute.
Both the patients and the controls were screened by renowned neuro-medicine clinical specialists (TKB and DPC) along with their team of trained neurologists at collaborating hospitals, Kolkata.
Collection of blood samples and genomic DNA preparation
Approximately 5 ml of peripheral blood samples was collected in K2 EDTA Becton Dickinson Vacutainer (6 ml) with written and informed consent from PD patients, their family members and from normal individuals as controls making sure about adequate understanding by donors. Genomic DNA was prepared from fresh whole blood by using conventional phenol-chloroform method.9 Genomic DNA was dissolved in TE (10 mM Tris-HCl, 0.1 mM EDTA, pH 8.0).
Polymerase chain reaction
PCR was carried out to amplify coding exons and adjacent flanking region in a total volume of 10.0 μl containing 40–100 ng genomic DNA, 0.4 μM of each primer, 0.2 mM of each dNTP, 0.5–1.5 mM of MgCl2 (as appropriate) and 0.5 unit of Taq polymerase (Invitrogen, Carlsbad, CA, USA) in a Thermocycler (GeneAmp-9700, PE Applied Biosystems, Foster City, CA, USA). PCR-amplified DNA fragments were analyzed on 2% agarose gel and visualized by ethidium bromide staining.
Mutation and polymorphism detection
DNA sequencing: the PCR products free of contaminating bands due to non-specific amplification were directly sequenced in forward and reverse direction using an ABI Prism 3730 DNA Analyzer (Applied Biosystems) and the Applied Biosystems BigDye Terminator Chemistry (Applied Biosystems). Nucleotide changes were detected by comparing the sequence obtained in chromatogram with the normal PARKIN gene sequence (GenBank Accession No. AB009973) using pair-wise BLAST10 and SeqScape software v2.5.
Gene expression
CSF from PD patients (n=10) and controls (n=7) were collected and total RNA was isolated using MagNA Pure LC 2.0 (Roche Diagnostics, Indianapolis, IN, USA). Reverse transcription was performed according to the manufacturers protocol to obtain cDNA (Applied Biosystems). Resulting templates were subjected to real-time PCR analyses using probe (probe #62) and primers (Forward: 5′-GACAGCAGGAAGGACTCACC-3′ and Reverse: 5′-AGGGGCCTTTGCAATACAC-3′) from the universal probe library of Roche Diagnostics, USA.
Gene dosage study by MLPA analysis
To determine the PARKIN gene dosage, MLPA assay was performed using the SALSA MLPA kit, P051-C1 (MRC Holland; Amsterdam, The Netherlands) according to the manufacturer’s protocol among 38 PD patients. The stratification of 38 DNA samples chosen for MLPA is as follows: (a) patient samples where variants for PARKIN gene were present (n=21) in which five samples underwent expression study. (b) Patient samples with no variants (n=17). The kit contained probes for the exons of PARK1, PARK2, PARK5, PARK6, PARK7 and PARK8 genes. The PCR fragments for exons 2–5, 8 and sequences were analyzed on a 3130 × 1 Genetic Analyzer and GeneMapper software v3.7 (Applied Biosystems). The relative peak height (RPH) for each exon from patients and normal individuals was compared. A ratio between 0.7 and 1.3 was considered as normal, 0.3 and 0.6 as heterozygous deletion. Absence of a peak (ratio 0.0) would indicate a homozygous deletion.
Statistical analysis
Baseline characteristics were compared between subjects with PD and controls by unpaired Student’s t-test. Data were expressed as mean (±s.d.). The gene counting method estimated allele frequencies for each genotype. To test the departure of allele frequency spectrum from the Hardy–Weinberg equilibrium, we employed χ2-test with one degree of freedom, using the HWSIM program. The association between the PD patients and single-nucleotide polymorphism (SNP) was examined according to dominant and recessive genetic models, and the P-value, odds ratio (OR) and 95% confidence intervals (CI) were calculated. All the statistical analyses were performed using the SPSS statistical software version 16.0 (SPSS, Chicago, IL, USA) for Windows. A P-value <0.05 (two-tailed) was considered statistical significant. In addition, patients were stratified by age at presentation and aggressiveness of disease to determine whether an association existed between genotype and age of onset and development of PD. The estimation of frequency of rearrangement mutation among 300 participants (PD patients=150; controls=150) was detected using Haplotype frequencies established by Arlequin v2.0.11 The Haploview 3.1212 with default settings was used to assess the Linkage disequilibrium (D′ and r2) between each pair of SNPs and also to define haploblocks.
Results
Evaluation of PARKIN variants in PD
The present study subjects of 150 PD patients ranged from 38 to 80 years (126 males and 24 females) with mean age of onset 52.3±8.17 years from unrelated families; Unified Parkinson disease Rating Scale score was 31.2±5.20; Hoehn and Yahr staging scale was 2.43±1.10. The mean age was 54.03±11.73 years (age range, 42–80 years; 130 males and 20 females) for all the 150 controls. The age of the controls and patients were similar, as were the sex ratios, demonstrating adequate matching (P=0.13 and P=0.23, respectively) (Table 1). Most of the patients reported an increase in tremor and gait imbalance during periods of stress. Only four patients were untreated, whereas others were under antiparkinsonian treatment. Family history for PD was found in 7.9% cases in the present study. Control samples were screened to identify nucleotide variants found in the patients by bidirectional sequencing.
The 150 PD patients recruited in this study were initially screened for mutations in DJ-1 and LRRK2 genes. None of these individuals harbored any pathogenic mutations in LRRK2 and DJ-1 genes.13, 14 On screening these patients for nucleotide variants in all the12 exons and flanking regions of Parkin gene, a total of 11 changes were identified (rs2075923(IVS2+25T>C), rs55777503(Gln34Arg), rs72480422(Asp280Asn), rs34424986(Arg275Trp), rs3765474(IVS7–35G>A), Val186Ile, IVS8+48C>T, rs199657839 (c.1101C>T(Arg334Cys)), rs1801474(Ser167Asn), rs1801582(Val380Leu), rs35125035(1547C>A (3′UTR); Figure 1), which include two novel changes and nine reported changes (Table 2). Among the sequence variants, five missense mutations (Table 2) were found only in the patients under study, but none in the 150 control individuals selected based on lack of any neurological symptoms. The allele and genotype frequencies were observed to be significantly different between PD and control groups for polymorphisms rs1801582 (X2genotype=8.106, P=0.017; X2allele=6.814, P=0.009) and rs1801474 (X2genotype=11.269, P=0.003; X2allele =7.399, P=0.0065). We did not find any LD among the SNPs, the low r2 for every pair of SNPs indicates that these SNPs cannot be tagged by each other.
Location of nucleotide variants in the Parkin gene. The Parkin gene, consisting of 12 exons, is shown schematically. The hatched regions show location of various motifs in the protein as indicated. All the mutations (boxed), single-nucleotide polymorphisms and rare variants are shown. A full color version of this figure is available at the Journal of Human Genetics journal online.
Among the variants, a novel mutation in Ex 5 involving an amino acid change from Val186Ile (GTT>ATT) was found in one patient, but absent in 300 chromosomes (Supplementary Figure 1a). This patient had an age of onset <50 years with tremor, rigidity on both sides of the body. In addition, common symptoms like pain, speech, impairment and impaired ocular movement was exhibited with the progression of the disease. This patient’s grandfather suffered from rest tremor and parkinsonian-like symptoms although he was not diagnosed as PD due to lack of treatment facilities. This patient’s occupation is agriculture and he was found to use pesticides at regular intervals. The PARKIN protein expression from CSF was almost 2.7-folds lower than the controls and other PD patients (PRK_507, Figure 3). In addition, enzyme assays of superoxide dismutase, catalase and glutathione peroxidase showed decrease levels in the plasma, thus leading to increased oxidative stress and generation of free radicals.15, 16, 17 The conservation of this amino acid suggests the importance of the residue and potential for functional aberration when mutated (Supplementary Figure 1b). FoldX stability analysis could not be carried out by SNPeffect software due to the lack of any reliable information regard the structural information of the PARKIN protein.
Arg334Cys was detected in a heterozygous state in a 58-year-old male patient. However, none of the above two variants was present in their offspring. The disease started with rest tremor and progression was observed fast for this patient. After 4 years of treatment, he developed drug-induced dyskinesia.
We found another change c1574 C>A in 3’UTR (rs35125035), not previously identified in Indian population, present in the heterozygous state of one patient. A novel intronic change 1VS8+48C>T was present almost equally in PD patients and controls (Supplementary Figure 2a) and was also found to be conserved among the other species (Supplementary Figure 2b).
The allele and genotype frequencies for polymorphisms and variants are shown in Table 2. The allele frequencies of some of the nucleotide variants identified in eastern Indian population are very low (0.43–0.006) and are rare variants. Other nucleotide variants identified in patients were not likely to be causal to PD as the changes were either located in introns (but introns in splice junctions).
In our study group, except for rs1801474 and rs3765474, both the control and PD groups were in the Hardy–Weinberg equilibrium. The Val380 allele (nt1239G, rs1801582) was significantly more frequent in the overall PD sample (P=0.012, OR=1.738, 95% CI, 1.21–2.697). The predisposing genotype was G/G (P=0.007, OR=2.02; 95% CI, 1.201–3.405; Table 2).
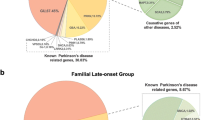
Similarly, for Ser167 allele (rs1801474), GG genotype also occurred more frequently in the Bengalee patients than in the control group (P=0.002, OR=3.355, 95% CI, 1.492–7.706; Table 2). Both the cSNPs in Parkin, within this cohort, were found to be significantly associated with PD independent of age of onset, sex and the presence of other polymorphisms. There was no linkage disequilibrium between the polymorphisms despite their location within the same gene, suggesting the existence of frequency recombination within the large introns of the Parkin gene. Analysis of the allele and genotype frequencies of different populations of the world included in the HAPMAP project suggested that Han Chinese in Beijing (CHB), CHD and Japanese in Tokyo (JPT) show similar profile that is remarkably distinct from Utah residents with ancestry from northern and western Europe (CEU), GIH, MEX, Yoruba in Ibadan, Nigeria (YRI) and Indian profile (Figure 2).
A comparison of the allele frequency distribution of (a) Ser167Asn (rs1801474), (b) Val380Leu (rs1801582), (c) rs2075923 and (d) 3765474 between the HapMap populations and the Bengalee population (present study) are shown. The numbers in the pie-charts represents the frequencies of the respective alleles of the SNPs.
The allele frequencies of the variants were calculated in the Bengalee population as well as in the world population (Hapmap data). This was done to check the utility of the SNP as a marker in our population as well as in the World population. The allele frequencies of the obtained SNPs have been illustrated by the pie-charts in Figure 2.
To check the informativeness of the SNPs in different world population, allele frequencies were compared. The four SNPs of Parkin gene evaluated in different populations are: rs1801474, rs1801582, rs2075923 and rs3765474 (Table 3). The observed heterozygosity of the SNPs was calculated in 150 normal individuals representing the control group. For the SNP rs1801582, the two mongoloid population CHB (Han Chinese from Beijing) and JPT (Japanese from Tokyo) showed similar profile, whereas our Indian population showed similar profile to the YRI, MEX and GIH population. It is observed that in populations like MEX (0.188), GIH (0.114), YRI (0.177) and data from present study (0.186) for rs1801474 is yielding a moderate heterozygosity (Table 3). Interestingly, rs2075923 shows the lowest heterozygosity value (0.18) for the Bengalee population (minor allele frequency =0.09) much in contrast to all other populations considered. rs3765474 reflects greater heterozygosity among all the populations studied with range varying from 0.386 to 0.534.
Analysis of Parkin protein expression showed reduced levels of the protein in CSF (Figure 2). Control_106 had no neurological symptom in his family and was healthy with an age of 54 years. PRK_CTRL147 (M, 77 years) and 08 (M, 71 years) were patients admitted in the collaborating hospitals in the neurosurgery department for diseases other than PD. Ctrl_466 (M, 43 years), 357 (M, 51years), 10 (F, 64 years) and 742 (M, 70 years) were individuals who were suffering from diseases other than any neurological disorders. PRK_68 (M, 56 years; H&Y stage: 1), 290 (M, 80 years; H&Y stage: 2), 125 (M, 59 years; H&Y stage: 2), 306 (F, 50 years H&Y stage: 1) did not harbor any variant in the PARKIN gene screened. PRK_139 (M, 61 years; H&Y stage: 3), 79 (M, 65 years; H&Y stage: 2), 34 (M, 49 years; H&Y stage: 3) had SNPs G/G genotype for Ser167, whereas PRK_02 (F, 53 years; H&Y stage: 3) for Val380, respectively. PRK_169 (M, 69 years; H&Y stage: 3) could not be genotyped due to some technical problems. PRK_34 had positive family history for PD (paternal uncle was detected with PD at the age of 72 years). PRK_02 and 125 was involved in farming and agriculture as occupation with the use of pesticides and insecticides for the past 10–12 years with rural living.
No deletion/duplication was found in 38 PD patients as analyzed by MLPA. The average ratio of parkin to β-globin was ~1.0 for all the patients.
Discussion
A wide variety of mutations in the Parkin gene have been repeatedly found in patients from many European, American, Asiatic and Chinese populations. The clinical signs showed 39% of the patients exhibited the three major cardinal symptoms of PD followed by 36% having tremor and bradykinesia in combination were present in patients at the time of examination. Among the other secondary symptoms, 25.51% have memory disturbances, 36.62% have micrographia, 34.56% have postural instability and 44% have sleep disturbances. Unilateral upper limb and lower limb tremor was found to be predominant symptom (53.03%) followed by bilateral rest tremor. Anxiety and stress increased the symptom. This is the first molecular analysis of the Parkin gene in an ethnically stratified Bengalee population of India. The ability to detect PRKN mutations depends on factors such as sample size, ethnic extraction, inclusion criteria for cases and the methods used for mutation detection. In our study, only six PD patients (4%) possessed PRKN mutation (Table 2), which is lower than that reported in several comparable studies, which report PRKN mutations to be present in between 10.4 and 18.0% of early-onset cases.18, 19, 20, 21 Five missense mutations identified in our patients are located in the important functional domain of Parkin, that is, one in UBL, another in UPD, two in RING1, whereas other in IBR region—thus likely to be involved in the impairment of Parkin function, thereby leading to decrease folds of the protein expression in CSF. Interestingly, none of the 300 chromosomes harbored the variants suggesting the potential role of these mutations, if any, in PD, require to be evaluated by functional analysis. The novel mutation (ex 5, GTT>ATT) is predicted to be ‘possibly damaging’ (PolyPhen, http://coot.embl.de/PolyPhen/; SNPs3D, Mutation taster) and the amino acid in this position is highly conserved through Macaca mulatta, Pan paniscus, Callithrix jacchus, Bos mutus, Sus scrofa, Rattus norvegicus and Canis familiaris. The mutation has not been found in previous other PD studies reported till date.
PD being a complex disease, the interplay of multiple genes and environmental factors is likely to contribute to the disease pathogenesis. The loss of Parkin-E3 enzyme function in the ubiquitin–proteasome pathway, resulting in the accumulation of PARKIN substrates in neurons, underlies the molecular basis of this disease by analyzing the PARKIN mutations.22, 23, 24 In comparing the clinical phenotype with the patients harboring PARKIN changes (Supplementary Table 1), all our patients demonstrated significant phenotypic heterogeneity with respect to age of onset, symmetrical presentations, disease duration and progression, disease severity, drug response although all had typical clinical and Parkinsonian features (rest tremor, rigidity, bradykinesia, gait imbalance, postural instability) All the characterized changes were observed in heterozygous condition.
As in previous studies among Asian,4, 24 we too found an association between PD and the allele or genotype frequency of the Ser167Asn polymorphism, unlike European population and white populations.18, 25 The frequency of this polymorphism appears to be highly dependent on ethnic origin (40% in Asian population).24 Increasing evidence indicates a role of heterozygous pathogenic mutations as a susceptibility factor for PD, although, meta-analysis strongly suggests that Parkin p.Ser167Asn variant is not associated with PD risk.26 Ser167Asn variant is insufficient to claim its general clinical importance for PD. Positron emission tomography imaging studies have reported a subclinical dopaminergic dysfunction in heterozygous Parkin mutation carriers.27 There is an increased frequency of heterozygous mutations in patients with PD compared with healthy controls,28 which is quite similar to our data. However, the literature is not consistent, as some studies report a similar frequency of heterozygous mutations in cases and in controls,29, 30 whereas, some studies support that heterozygous Parkin point mutations are not associated with PD.31
In this study, we detected a positive association between PD and homozygosity for the Val380 allele, similar to studies from India4 and European population; although no association with this allele was found in two Asian PD population in which the frequency of the variant allele was low (4–5%).24, 32 Our current findings support a relationship between the presence of the Val allele and increased risk for the presence of PD as proven in meta-analysis of this SNP by Zang et al. 33 However, the presence of the Val allele is neither necessary nor sufficient for the development of PD and the mechanism(s) through which this risk allele may exert its effects or conversely, the presence of the Leu allele may protect against the development of PD, are uncertain. The Parkin p.Val380Leu polymorphism (the presence of Leu allele at codon 380 in Parkin) is located in the RING-IBR-RING domain of Parkin which suggests it could act by affecting the binding of substrates or protein ubiquitin-conjugating enzymes.22, 23 In addition, Parkin p.Val380Leu polymorphism might alter interactions with environmental factors acting as a disease modifier.34 For the Val380Leu and Ser167Asn polymorphism, presence of the most frequent alleles (found in 77 and 90% of controls, respectively; Figure 2) increased the risk of PD, suggesting that the rare alleles might be protective which is at par with earlier findings of Lucking et al.18 and Biswas et al.,4 contrary to Martinez et al.35
Gene expression quantification assays, showed a decreased expression in PARKIN gene in PD patients, further validating the findings. Previous studies had investigated the PARK2 mutations without any emphasis on quantitation. Our results showed various folds of alterations of Parkin protein expression in PD patients’ CSF (Figure 3). Even patients without mutations exhibited significantly reduced Parkin protein expression. The multifactorial pathological conditions such as PD have been reported to involve oxidative stress and aging, which may have an important role in addition to the genetic factors. It is possible that these factors modify the Parkin translational machinery in such patients independent of Park2 mutations. Although studies from Indian population detected deletions in exons 3–4 (two siblings in 138 patients; 1.449%)4 and exons 8–9 (one in 102 patients; 0.98%),3 it can be concluded to the occurrence of low frequency of deletions in the cohorts studied, similar to our findings, quite contrary to heterozygous exon rearrangements observed in 9.2% of North Indian population while absence of homozygous exonic deletions.5
Some possible limitations of our study should be acknowledged. First, selection bias in the study might have affected our results, although the genotype distribution of patients and controls in our study was compatible with the Hardy–Weinberg expectations. Second, our sample size was not big enough and our study was performed in a local Bengalee population. The study should be extrapolated to other regions and ethnic groups cautiously. However, this internally consistent pilot study has provided valuable information for future studies in this area.
In conclusion, our results demonstrate the occurrence of Parkin mutation in 4% (6/150) of our PD patient pool supported with decreased folds of expression of CSF PARKIN protein. Our data clearly indicate that homozygous Ser167 and Val380 are significantly associated with the disease suggesting that the Parkin polymorphisms are risk factors for PD, and their frequency is greatly influenced by ethnic origin.
References
Greenamyre, J. T . & Hastings, T. G . Parkinson’s–divergent causes, convergent mechanisms. Science 304, 1120–1122 (2004).
Tang, S ., Zhang, Z ., Kavitha, G ., Tan, E. K . & Ng, S. K . MDPD, an integrated genetic information resource for Parkinson’s disease. Nucleic Acids Res. 37, 852–862 (2009).
Madegowda, R. H ., Kishore, A . & Anand, A . Mutational screening of the Parkin gene among South Indians with early onset Parkinson’s disease. J. Neurol. Neurosurg. Psychiatry 76, 1588–1590 (2005).
Biswas, A ., Gupta, A ., Naiya, T ., Das, G ., Neogi, R ., Datta, S . et al. Molecular pathogenesis of Parkinson’s disease, Identification of mutations in the Parkin gene in Indian patients. Parkinsonism Relat. Disord. 12, 420–426 (2006).
Chaudhary, S ., Behari, M ., Dihana, M ., Swaminath, P. V ., Govindappa, S. T ., Jayaram, S . et al. Parkin mutations in familial and sporadic Parkinson’s disease among Indians. Parkinsonism Relat. Disord. 12, 239–245 (2006).
Fahn, S ., Elton, R.L ., members of the UPDRS Development Committee. in: Recent developments in Parkinson's Disease, Vol. 2 (eds Fahn, S ., Marsden, C. D ., Calne, D. B . & Goldstein, M .) 153–163 (Macmillan Health Care Information, Florham Park, NJ, 1987).
Hoehn, M. M . & Yahr, M. D . Parkinsonism: onset, progression, and mortality. Neurology 17, 427–442 (1967).
Folstein, M. F ., Folstein, S . & Mchugh, P. R . Mini-Mental State: a practical method for grading the mental state of patients for clinicians. J. Psychiatr. Res. 12, 189–198 (1975).
Sambrook, J . & Russel, D. W . (eds). Molecular Cloning: a Laboratory Manual 6.23–6.27, 3rd edn (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY, 2001).
Tatusova, T . & Madden, T . BLAST 2 sequences a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174, 247–250 (1999).
Schneider, S ., Roessli, D . & Escoffier, L . Genetics and Biometry Laboratory, (University of Geneva: Switzerland, 2000).
Barrett, J. C ., Fry, B ., Maller, J . & Daly, M. J . Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005).
Sanyal, J ., Sarkar, B. N ., Ojha, S ., Banerjee, T. K ., Ray, B. C . & Rao, V. R . Absence of commonly reported leucine-rich repeat kinase 2 mutations in Eastern Indian Parkinson’s disease patients. Genet. Test. Mol. Biomarkers 14, 691–694 (2010).
Sanyal, J ., Sarkar, B. N ., Banerjee, T. K ., Mukherjee, S. C ., Ray, B. C . & Rao, V. R . Evaluating intra-genetic variants of DJ-1 among Parkinson’s disease patients of Eastern India. Neurol. Res. 33, 349–353 (2011).
Sanyal, J ., Bandyopadhyay, S. K ., Banerjee, T. K ., Mukherjee, S. C ., Chakraborty, D. P ., Ray, B. C . et al. Plasma levels of lipid peroxides in patients with Parkinson’s disease. Eur. Rev. Med. Pharmacol. Sci. 13, 129–132 (2009).
Sanyal, J ., Sarkar, B ., Banerjee, T. K ., Mukherjee, S. C ., Ray, B. C . & Rao, V. R . Plasma level of nitrates in patients with Parkinson’s disease in West Bengal. Neurol. Asia 15, 55–59 (2010).
Sanyal, J ., Sarkar, B ., Banerjee, T. K ., Mukherjee, S. C ., Ray, B. C . & Rao, V. R . Peripheral markers for oxidative stress in Parkinson’s disease patients of Eastern India. Neurochem. J. 5, 146–149 (2011).
Lucking, C. B ., Durr, A ., Bonifati, V ., Vaughan, J ., De, Michele, G ., Gasser, T . et al. Association between early-onset Parkinson’s disease and mutations in the parkin gene. N. Engl. J. Med. 342, 1560–1567 (2000).
Periquet, M ., Latouche, M ., Lohmann, E ., Rawal, N ., De Michele, G ., Ricard, S . et al. Parkin mutations are frequent in patients with isolated early-onset Parkinsonism. Brain 126, 1271–1278 (2003).
Sun, M ., Latourelle, J. C ., Wooten, G. F ., Lew, M. F ., Klein, C ., Shill, H. A . et al. Influence of heterozygosity for parkin mutation on onset age in familial Parkinson disease: the Gene PD study. Arch. Neurol. 63, 826–832 (2006).
Hertz, J. M ., Ostergaard, K ., Juncker, I ., Pedersen, S ., Romstad, A ., Moller, L. B . et al. Low frequency of parkin, tyrosine hydroxylase, and GTP cyclohydrolaseI gene mutations in a Danish population of early-onset Parkinson’s Disease. Eur. J. Neurol. 13, 385–390 (2006).
Shimura, H ., Schlossmacher, M. G ., Hattori, N ., Frosch, M. P ., Trockenbacher, A ., Schneider, R . et al. Ubiquitination of a new form of alpha-synuclein by Parkin from human brain, implications for Parkinson’s disease. Science 293, 263–269 (2001).
Imai, Y ., Soda, M . & Takahashi, R . Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J. Biol. Chem. 275, 35661–35664 (2000).
Chung, K. K ., Zhang, Y ., Lim, K. L ., Tanaka, Y ., Huang, H ., Gao, J . et al. Parkin ubiquitinates the a-synuclein interacting protein, synphilin-1, implications for Lewy-body formation in Parkinson disease. Nat. Med. 7, 1144–1150 (2001).
Eerola, J ., Launes, J ., Hellstrom, O ., Tienari, P. J . & Apolipoprotein, E (APOE), PARKIN and catechol-o-methyltransferase (COMT) genes and susceptibility to sporadic Parkinson’s disease in Finland. Neurosci. Lett. 330, 296–298 (2002).
Zhang, Y ., Wang, Z. Z . & Sun, H. M . Lack of association between p.Ser167Asn Variant of Parkin and Parkinson's disease, A meta-analysis of 15 studies involving 2,280 cases and 2,459 controls. Am. J. Med. Genet. 159B, 38–47 (2012).
Khan, N. L ., Scherfler, C ., Graham, E ., Bhatia, K. P ., Quinn, N ., Lees, A. J . et al. Dopaminergic dysfunction in unrelated, asymptomatic carriers of a single Parkin mutation. Neurology 64, 134–136 (2005).
Klein, C ., Lohmann-Hedrich, K ., Rogaeva, E ., Schlossmacher, M. G . & Lang, A. E . Deciphering the role of heterozygous mutations in genes associated with parkinsonism. Lancet Neurol. 6, 652–662 (2007).
Lincoln, S. J ., Maraganore, D. M ., Lesnick, T. G ., Bounds, R ., de Andrade, M ., Bower, J. H . et al. Parkin variants in North American Parkinson’s disease, cases and controls. Mov. Disord 18, 1306–1311 (2003).
Kay, D. M ., Moran, D ., Moses, L ., Poorkaj, P ., Zabetian, C. P ., Nutt, J . et al. Heterozygous parkin point mutations are as common in control subjects as in Parkinson’s patients. Ann. Neurol. 61, 47–54 (2007).
Chien, H. F ., Rohe, C. F ., Costa, M. D ., Breedveld, G. J ., Oostra, B. A ., Barbosa, E. R . et al. Early-onset Parkinson’s disease caused by a novel Parkin mutation in a genetic isolate from north-eastern Brazil. Neurogenetics 7, 13–19 (2006).
Hu, C. J ., Sung, S. M ., Liu, H. C ., Lee, C. C ., Tsai, C. H . & Chang, J. G . Polymorphisms of the parkin gene in sporadic Parkinson's disease among Chinese in Taiwan. Eur. Neurol. 44, 90–93 (2000).
Zhang, Y ., Wang, Z. Z . & Sun, H. M . A Meta-Analysis of the Relationship of the Parkin p.Val380Leu Polymorphism to Parkinson’s disease. Am. J. Med. Genet. 162B, 235–244 (2013).
Aguiar, P. C ., Lessa, P. S ., Godeiro, C. J ., Barsottini, O ., Felicio, A. C ., Borges, V . et al. Genetic and environmental findings in early-onset Parkinson’s disease Brazilian patients. Mov. Disord 23, 1228–1233 (2008).
Martinez, H. R ., González-González, H ., Cantú-Martínez, L ., Rangel-Guerra, R ., Hernández-Castillo, C. D . & Vergara-Saavedra, J. J. J . et al. PARKIN-coding polymorphisms are not associated with Parkinson's disease in a population from northeastern Mexico. Neurosci. Lett. 468, 264–266 (2010).
Acknowledgements
This research work was supported by the grants from the Anthropological Survey of India, Ministry of Culture, Government of India. We are thankful to the PD patients and control subjects for voluntarily taking part in this research work and donating their blood samples. We also thank Mitali Maity (nurse in NNC) for helping in the collection of blood samples from the OPD of NNC. JS is thankful to CSIR for providing the Research Associate fellowship (CSIR RA code ID: 9/45(1283)/2013 EMR-I).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Sanyal, J., Jana, A., Ghosh, E. et al. Evaluation of PARKIN gene variants in West Bengal Parkinson’s disease patients. J Hum Genet 60, 485–492 (2015). https://doi.org/10.1038/jhg.2015.49
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2015.49
This article is cited by
-
PARK2 and PARK7 Gene Polymorphisms as Risk Factors Associated with Serum Element Concentrations and Clinical Symptoms of Parkinson’s Disease
Cellular and Molecular Neurobiology (2020)