Abstract
Mercury (Hg) is an anthropogenic pollutant that is toxic to wildlife and humans, but the response of remote ecosystems to globally distributed Hg is elusive. Here, we use DNA extracted from a dated sediment core to infer the response of microbes to historical Hg delivery. We observe a significant association between the mercuric reductase gene (merA) phylogeny and the timing of Hg deposition. Using relaxed molecular clock models, we show a significant increase in the scaled effective population size of the merA gene beginning ~200 years ago, coinciding with the Industrial Revolution and a coincident strong signal for positive selection acting on residues in the terminal region of the mercuric reductase. This rapid evolutionary response of microbes to changes in the delivery of anthropogenic Hg indicates that microbial genomes record ecosystem response to pollutant deposition in remote regions.
Similar content being viewed by others
Introduction
Mercury (Hg) is a contaminant globally distributed via the atmosphere that not only persists in the environment but is also biomagnified through aquatic food webs, particularly in its methylated form, monomethylmercury (Morel et al., 1998). Both Hg cycling and bioaccumulation depend on microbial processes (Parks et al., 2013). Human activities associated with industrialisation have exacerbated Hg contamination, but we currently have no good historical records of how freshwater ecosystems responded to the delivery of anthropogenic Hg. Indeed, although freshwater fish Hg levels respond quickly to atmospheric Hg deposition (Harris et al., 2007), they leave no historical records except for the very few that can be found in museum specimens (Miller et al., 1972; Drevnick et al., 2007).
Because of the complexity of its biogeochemical cycle, time lags in the response of many freshwater ecosystems to changes in atmospheric Hg levels can be expected (Brigham et al., 2014), hampering our ability to manage Hg contamination. This inability to properly describe ecological linkages between historical changes in Hg deposition and ecosystem response has been identified as a major research gap in the latest United Nation Environment Program Global Mercury Assessment (UNEP, 2013).
Recent advances in genotyping have shown that ancient DNA extracted from lake sediments or permafrost can provide information on the genome of organisms living in those environments (Coolen and Overmann, 2007; Coolen et al., 2008; Boere et al., 2011; D'costa et al., 2011; Takano et al., 2012; Lejzerowicz et al., 2013) and microbial DNA can be used to reconstruct past environments (Inagaki et al., 2005; Lyra et al., 2013), but we lack information on how microbes responded to changes in historical environmental pollution. Microbial community structure changes when exposed to metal concentration gradients (Turpeinen et al., 2004; Gough and Stahl, 2011) and this structuring could possibly serve as an index for establishing contemporary ecosystem responses to contaminant exposure. However, steep geochemical gradients typically develop within the very first few millimetres of sediments, strongly affecting redox conditions and re-shaping the structure of live microbial communities (Bertics and Ziebis, 2009; Stoeva et al., 2014). This structuring makes historical reconstruction of bacterial and archaeal communities using solely 16S rRNA markers often unreliable, as microbes will respond to geochemical changes observed during diagenesis. In the case of Hg, we can circumvent this issue and develop a reliable marker of historical exposure to Hg deposition, by focusing our analysis on genes of the mer-operon, encoding for microbial Hg detoxification (via Hg scavenging, uptake and reduction). These genes are ideal candidates in offering molecular insights into historical exposure to Hg because they are (i) specific to Hg detoxification (Barkay et al., 2003) and (ii) ubiquitous in the environment (Osborn et al., 1997; Barkay et al., 2010) and easily mobilised between microbial populations via horizontal gene transfer (Barkay et al., 2003). As such, we do not expect taxonomic discrimination of microbes based on their potential to detoxify Hg. Should the microbial community structure change, genes involved in Hg resistance should not be segregated based on species but rather maintained in the community based on whether they provide increased fitness in the environment considered.
Our study focused on the Hudson Bay Lowlands of subarctic Canada, as it contains one of the last Arctic cold refugia only recently lost to global warming (Rühland et al., 2013). In this unique environment, remote from direct human disturbances, we extracted nucleotide sequences encoding for Hg detoxification from dated lake sediments to test the hypothesis that these ancient DNA sequences reflect both the timing of sediment deposition and historical Hg exposure. Our results show a significant association between mercuric reductase gene (merA) phylogeny and the timing of Hg deposition and a significant increase in the scaled effective population size of the prokaryotes involved in Hg detoxification, beginning ~200 years ago, which coincides with the Industrial Revolution. Further analyses show that this event is driven by positive selection. Our approach offers novel insights into the effects of Hg emissions in remote regions, providing a new historical perspective on the evolutionary response of ecosystems to contaminants released since the onset of the Industrial Revolution.
Materials and methods
Sampling site and sediment collection
Sediment cores were collected from Aquatuk, Hawley and North Raft lakes in August 2010 in the Hudson Bay Lowlands of Ontario, Canada, which range in longitude and latitude from 53° to 55° N and 84° to 85° W, respectively. Aquatuk is a mesotrophic lake and Hawley and North Raft are oligotrophic lakes and all three are located on the Precambrian bedrock outcrop of the Sutton Ridges, although their relatively high pH (ca. 8.0) and ionic strength ([Ca2+] ca. 30 mg l−1) reflect the drainage from the surrounding calcareous terrain of the Hudson Bay Lowlands (Gunn and Snucins, 2010; Paterson et al., 2014). Sediment cores were taken using a Glew gravity corer (Glew, 1989). The sediment cores were sectioned into 0.5 cm intervals over the first 10 cm (20 samples) and every 2 cm for the remaining fraction of the core. Upon collection, core samples were divided in two; half of the interval was put into a 50 ml polypropylene centrifuge tube with 10 ml of RNAlater and shaken to allow the solution to properly penetrate the sample, which was dedicated to molecular analyses. The other half of the interval was placed into a Whirl-Pak bag (Nasco, Fort Atkinson, WI, USA) and dedicated to Hg and radiometric analyses. All samples were kept at 4 °C and in the dark until frozen at −20 °C. All containers used for sampling were sterile, extruding instruments were rinsed with water between each layer and non-powdered gloves were worn at all times during sampling and processing of the samples. Full characterisation of all lakes used in this study is presented elsewhere (Brazeau et al., 2013).
Mercury analyses
Frozen sediments were transferred from Whirl-Pak bags into 50 ml sterile high-density polypropylene Falcon tubes (Corning, Tewksbury, MA, USA). Samples were then lyophilised for a period of 72 h under a vacuum of 5 atm and then homogenised. Total Hg in sediments was analysed with Nippon Instruments Corporation’s Mercury SP-3D Analyser (CV-AAS) (Osaka, Japan) by thermal decomposition with gold trap amalgamation and cold vapour atomic absorption method (UOP Method 938-00, detection limit of 0.01 ng Hg and range up to 1000 ng Hg). The instrument was calibrated with Mercury Reference Solution 1000 ppm ±1% (Fisher CSM114-100, Waltham, MA, USA) and MESS-3 (91±9 ng g−1, National Research Council of Canada) was used as reference material. Blanks were performed as suggested by the manufacturer.
Radiometric dating
Sediment cores were radiometrically dated using gamma (γ) spectrometry and analysed for the activity of 210Pb, 137Cs and 226Ra in an Ortec germanium crystal well detector (DSPec, Ortec, model # GWL-120230, Oak Ridge, TN, USA) following the method by Appleby (Appleby, 2001). Analysis of 210Pb was performed on 14–18 selected depth intervals in the sediment cores to determine the sediment age, and the sediment accumulation rate. Samples were lyophilised, homogenised and hermetically sealed and left to reach secular equilibrium for a minimum of 21 days before being counted for 23 h (82 800 s). The resulting spectrum files showed 210Pb activity with a peak at 46.5 keV, and 137Cs at 662 keV. 226Ra activity was determined by γ-ray emissions of its daughter isotope 214Pb, resulting in peaks at 295 and 352 keV. Long-term sedimentation rates were determined for each core using the Constant Rate of Supply model from Appleby and Oldfield (1978). 137Cs activity (from atmospheric fallout of nuclear weapons, peaking in 1963) was measured to verify 210Pb dates. 210Pb results indicated that sediment mixing was not happening in these cores. Physical mixing typically appears as uniform 210Pb activity to the bottom of the mixed layer. These data do not support movement of particles (and therefore particle-bound) bacteria throughout the profiles.
Sample preparation for DNA work
Prior to subsampling for DNA extractions, sediment samples were thawed and centrifuged at 4000 r.p.m. for 10 min and the supernatant was discarded. For DNA extraction, a subsample of 0.5 g was placed into a sterile 2 ml Eppendorf tube. We used a sterile buffer modified from Zhou et al. (1996) to wash the sediment to remove contaminants such as humic substances and divalent cations that can be co-extracted with nucleic acids and inhibit subsequent downstream applications. The buffer consisted of 10 mM EDTA, 50 mM Tris-HCl and 50 mM Na2HPO4·7H2O at pH 8.0. We added three parts buffer for one part sediment, then samples were vortexed at maximum speed for 30 s, centrifuged at 4000 r.p.m. for 3 min and the supernatant was discarded. The washing procedure was repeated until the supernatant was clear (maximum of five washes). The washed sediment was immediately used for DNA extraction. All handling and processing of the samples occurred in a dedicated stainless-steel biological safety cabinet equipped with a HEPA 100 filter. All work surfaces were thoroughly washed with ethanol (70%), bleach (10%) and treated with UV radiations before and after each use. Non-powdered gloves were worn at all time.
DNA extraction and quantification
DNA extracted from any depths in sediments can reflect: (i) dormant or resting organisms (Hirsch et al., 2010); (ii) free DNA molecules adsorbed onto sediment particles (Haile et al., 2007); or (iii) live microbes. Bacterial DNA extraction from the washed sediment (0.25 g) was performed using a PowerSoil DNA Isolation Kit (MoBio, Carlsbad, CA, USA) according to the manufacturer’s guidelines. DNA extraction was performed on sediment from five depth intervals of each core. The extracted DNA was quantified using an Invitrogen Quant-iT dsDNA High-Sensitivity Assay Kit (Invitrogen, San Diego, CA, USA) according to the manufacturer’s protocol. DNA extracts were stored at −20 °C until further use. Note that the poor quality and yield of DNA obtained for North Raft Lake at depths of 10 and 22 cm did not allow for subsequent analyses to be performed.
Quantitative PCR (qPCR)
qPCR reactions were performed using the commercial enzyme kit SsoFast EvaGreen Supermix (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer's recommendations. Final per reaction reagent concentrations were: 1 × SsoFast EvaGreen Supermix and 0.2 μM Forward/Reverse primers (Supplementary Table S1). Samples and associated assay standards were assayed in triplicate with each replicate having a final volume of 20 μl. Samples and associated assay standards were run using the Eco Real-Time PCR System (Illumina Inc., San Diego, CA, USA). Machine user interface software used was the Eco Real-Time PCR system qPCR Software v2.0.6.0 (Illumina Inc). All primers used in this study are presented in Supplementary Table S1.
qPCR standards were created using PCR amplicon products derived from laboratory pure cultures known to house the gene of interest (Pseudomonas aeruginosa for mer-operon genes and Escherichia coli for glnA gene). The control gene, glnA, encodes the glutamine synthetase, which, as a core housekeeping bacterial marker, is expected to reflect overall bacterial abundance (Lauro et al., 2011). After the successful amplification of the partial gene segment of interest, the amplicons were cloned within the vector pGEM-T Easy Plasmid (Promega, Madison, WI, USA) and used to transform a competent stock of laboratory culture (E. coli strain XL10). The transformed culture was grown overnight and the vector plasmid housing the amplicon of the gene of interest was extracted using the Wizard Plus SV Minipreps DNA Purification System (Promega). Extracts were stored at −20 °C until further use. Concentration values for standard copy number were entered into the Eco Real-Time PCR system qPCR Software based on the volume (μl) inputs in each reaction vessel. Tenfold-based dilution series of the standard plasmid stock were generated to create a dilution curve for the qPCR standards. Standard quantities used in the generation of standard curves ranged from 10 copies to 107 copies of gene specific amplicon. qPCR results and standard curve data are presented in Supplementary Table S2.
Conventional PCR
PCR reactions were performed using the Eppendorf MasterCycler ProS (Eppendorf Canada, Mississauga, ON, Canada) and the Biometra T Professional Basic Gradient (Biometra, Goettingen, Germany). Commercial enzyme Platinum Taq Polymerase (Invitrogen) was used to generate PCR amplicons. Results of conventional PCR assays (for example, a clean, unique, distinct band on a gel) were used to ensure assay specificity and quality prior to the application of PCR protocols in quantification assays (qPCR) (QA/QC).
PCR reactions were carried out according to the manufacturer’s recommendations with final per reaction reagent concentrations being: 1 × PCR buffer, 2.5 mM MgCl2, 0.2 μM forward/reverse primers (Supplementary Table S1), 0.2 mM dNTPs, 0.625 U Platinum Taq Polymerase. For the determination of optimal PCR conditions in generating specific amplicon products with maximised yields, MgCl2 concentrations varied from 1.5 to 3.5 mM; optimal Mg2+ concentration was chosen based on the presence of a distinct and unique band of the expected size. Once generated, amplicons were separated by electrophoresis, and visualised under UV light. Genes targeted in PCR assays and their associated PCR primers with respective information on cycling conditions are presented in Supplementary Table S1.
MerA and rpoB gene clone libraries
Clone libraries were constructed using the Aquatuk Lake sediment core. This core was chosen for this more focused work as it provides consistent DNA quality throughout the profile (for example, we could not extract good quality DNA from deep sediment samples in North Raft Lake; Figure 1c) and for which total mercury concentration profiles did not exhibit perturbation (as observed for Hawley Lake, Figure 1b). We amplified merA genes using a nested approach; first PCR round using primer pair Nlf F and Nlf R, amplifying a ca. 1200 bp fragment, and a second round of amplification using primer pair Nsf F and Nsf R, amplifying a diagnostic 300 bp fragment. Amplicons were gel purified with a Qiagen QiaexII Purification Kit (Qiagen cat#20021, Valencia, CA, USA) and cloned using the StrataClone PCR Cloning Kit (StrataClone PCR Clonning Kit Cat#240205, Agilent, Santa Clara, CA, USA), as per the manufacturer’s instructions. RpoB gene fragment (640 bp) was amplified using one round of PCR. One DNA library of 96 clones was constructed per depth for each gene of interest (surface, middle and deep). All clones were sequenced by Beckman Coulter Genomics (Danvers, MA, USA). Genes targeted in PCR assays and their associated PCR primers with respective information on cycling conditions are presented in Supplementary Table S1.
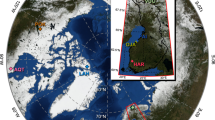
Depth profiles within sediment cores of three lakes. Depth profiles of total Hg concentrations, merA, merT, merP and glnA gene copy numbers collected in Aquatuk (a), Hawley (b) and North Raft (c) Lakes in Northern Ontario. Coloured boxes in panel (a) (red: 2010, green: ca. 1900 and blue: prior to 1800) correspond to the three depths chosen for rpoB and merA gene fragments sequencing.
Sequence analyses
The workflow for processing sequence information for phylogenic investigation was as follows: Vector sequences were removed from the merA and rpoB sequences using the vector annotation tool in Geneious v5.4 (Maidak et al., 2001). Primers from both merA and rpoB sequences were removed using Geneious’ primer annotation tool. An alignment of 209 known merA sequences retrieved from the NCBI database was built; this alignment was used to eliminate chimeric sequences. Chimeric sequences were identified using uchime (Edgar et al., 2011) in MOTHUR (Schloss et al., 2009). Sequences identified as chimeric were discarded. A total of 236 sequences were used for merA (90 at the surface, 92 at the middle and 54 at depth). A total of 269 sequences was used for rpoB (89 at the surface, 86 at the middle and 88 at depth). MerA and rpoB sequences were aligned based on the amino acid sequence using TranslatorX (Abascal et al., 2010). The model of evolution most appropriate to each of our datasets was determined using jModelTest v.0.1.1 with the Akaike Information Criterion (Posada, 2008). Distance matrices were constructed by building a phylogenetic tree in FastTree (Price et al., 2009) under the appropriate model of evolution. Sequence data reported in this paper can be found in GenBank under accession numbers JX308316:JX308551 for merA and KM204402:KM204658 for rpoB.
Statistical analyses
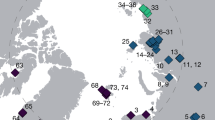
Analyses of the molecular data were performed with BEAST (Drummond and Rambaut, 2007) with the models described in Figure 2. Markov chain Monte Carlo samplers were run for 500 million generations with a thinning of 5000 to decorrelate samples; chains were run in duplicate to assess convergence on the target distributions. Model selection was based on the path sampling method (Xie et al., 2011) implemented in BEAST ver.1.7.2 to estimate marginal log-likelihoods and rank models. Ancestral demographic reconstructions were based either on a constant population size coalescent model or on a coalescent skyline model (Drummond et al., 2005) with 10 breakpoints and piecewise constant spline regressions (Supplementary Text).
Model selection and paleo-demographic reconstructions. Models are ranked from top to bottom by increasing estimates of marginal log probabilities of each model, based on two runs; error bars: 2 standard deviations. Compared models are: All.equal.const: all three depths are contemporaneous (equal prior dates) with constant population size (demography); Two.dates.const: two prior dates (0 for superficial samples; 500 y for all other samples), constant demography; Three.dates: three prior dates (0 for superficial samples; 125 y for intermediate depth; trailing number is prior age for deepest samples); sky: skyline demographic model. Bayesian skyline demographic reconstruction under the best model is shown in the inset; dashed vertical line in inset shows timing of the Industrial Revolution, ca. 200 years ago.
The association between phylogeny and sampling depths was assessed with BaTS (Parker et al., 2008) using 103 replicates on a subsample of 104 trees sampled from the posterior distributions for merA and rpoB under the model with largest marginal log-likelihood. Trees subsamples were obtained with a custom R script (Supplementary Text).
Sites potentially under Hg selection were identified with the DataMonkey pipeline (Delport et al., 2010) under a model of episodic positive evolution (Murrell et al., 2012); this is a mixed effects model where nonsynonymous to synonymous rates vary among both amino acid sites (the fixed effect model) and branches (the random effect model); a Likelihood Ratio Test was then used to compare a model where positive selection is permitted vs one where it is not (the null model) and hence infer site-wise selection; because each site is tested multiple times, P-values need to be corrected for False Discovery Rate by means of q-values (Storey and Tibshirani, 2003). The frequency of the mutations under selection, as present in GenBank, was obtained by running tBLASTn searches; its phylogenetic distribution was estimated by aligning the MerA sequences resulting from the tBLASTn searches at the protein level with Muscle (Edgar, 2004), and estimating a maximum likelihood tree under GTR + Γ with FastTree. Three-dimensional models of the partial MerA protein were estimated with SwissModel (Bordoli and Schwede, 2012). Protein domains were identified from Pfam (Finn et al., 2014).
Results and discussion
Sediment cores from temperate and northern regions provide a record of recent changes in Hg deposition and suggest that modern Hg deposition in the northern hemisphere is two to four times greater than pre-industrial background values (Amos et al., 2013). First, to determine whether microbial populations responded to an increase in Hg delivery with an increase in the copy numbers of the operon, we quantified the copy numbers of three mer-operon genes, merP (encoding Hg scavenging), merT (encoding Hg transport) and merA (encoding Hg reduction), and of a control housekeeping gene (glnA) over time, along with total Hg levels recorded in the sediment cores of three lakes located in the same area (Brazeau et al., 2013). Here, while total mercury increased twofold from old to recently deposited sediments (Figures 1a–c), mirroring the impact of global anthropogenic Hg distribution and deposition in this region, we found that copy numbers of all three genes of the mer-operon declined ca. 10-fold; glnA gene copy numbers (our control gene) remained constant throughout most of the profile, changing only at the surface, likely reflecting bacterial dynamics at the sediment/water interface where delivery of fresh nutrients occurs. This negative relationship between mer-gene copy numbers and total mercury for all three lakes (whether it is normalised per ng of DNA extracted or per copy number of glnA) through time implies either (i) that our quantitative approach could not detect sequences most abundant at the surface (for example, owing to sequence changes in primer binding sites or matrix effects) or (ii) that microorganisms cope with Hg selective pressure by means other than a quantitative change in detoxifying enzymes. An increase in Hg-driven selective pressure may have led to microbes producing mer-operon encoded enzymes with an increased activity. We therefore predicted that changes in the delivery of Hg would be recorded in the sequences of genes involved in Hg detoxification and stored in sediments.
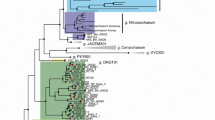
We tested this prediction by developing clone libraries for two genes: merA, our Hg-detoxifying marker, and the β-subunit of the bacterial RNA-polymerase (rpoB), a second housekeeping gene that is not expected to have evolved in response to changes in Hg delivery to aquatic ecosystems. We performed this detailed analysis on the sediment core of Aquatuk Lake for three depths, corresponding to sediments deposited recently (2010, year of sampling), ca. 1885 (shortly after the onset of industrialisation; Amos et al., 2013) and from the bottom of the core estimated to predate the 1800s (that is, pre-industrial times) based on 210Pb core dating and known sedimentation rates for this area (Rühland et al., 2013). We found a significant phylogenetic structuring of sampled sequences with sampling depth in the case of merA (P<10−3) but not in the case of rpoB (P=0.15) (Figure 3; see also Supplementary Figures S1 and S2). Note that calculations of merA genetic diversity via the Shannon index (S) show that diversity decreased from bottom (S=1.23±0.14) to top (S=1.03±0.19), but not significantly (t=0.85, df=176, P=0.40). This clustering pattern reflects the ancestral nature of merA in deeper sediments and suggests its rapid evolution following changes in the delivery of anthropogenic Hg. Because the Hudson Bay Lowlands environment only recently experienced warming (that is, mid-1990s) as shown using diatom paleo-indicators (Rühland et al., 2013), any changes in merA gene sequences observed prior to 25 years ago were likely not driven by warming but presumably by Hg delivery to lake sediments; indeed, this unique area offered the possibility to explore the dynamics of genes involved in Hg detoxification without the pressure of possible confounding factors exerted by climate change.
MerA and rpoB gene phylogenies. Example of phylogenetic reconstructions, showing the nonlinear relationship between sediment core depth and divergence times for merA (a) and rpoB (b) genes in Aquatuk lake sediments. The evolution of merA sequences is highly contingent: only a small fraction of sequences persist through time. Coloured lines (red: 2010, green: ca. 1900 and blue: prior to 1800) correspond to the three depths chosen for rpoB and merA gene fragments sequencing.
Expression of the mer-operon is tightly regulated by intracellular Hg levels (Barkay et al., 2003) and studies have shown that merA was expressed in environments far removed from direct Hg contamination sources exhibiting trace levels of Hg, such as Canadian boreal lakes (Siciliano et al., 2002) or in the Canadian High Arctic (Poulain et al., 2007). It is therefore reasonable to propose that a twofold increase in Hg levels observed in our sedimentary record (Figures 1a–c) may have selected microbes exhibiting an increased activity of their mercuric reductase (MerA). To address this question, we ran a codon model that detects episodes of positive selection acting at sites that are a priori unknown (Murrell et al., 2012); this approach identified one site with strong statistical signal (Supplementary Table S3: A375L; P<0.0001; q-value=0.01) that is located on all branches leading to surface sequences except for three sequences (Supplementary Figure S3). Microbial adaptation to Hg stress is likely to be complex and will rely on the generation of genetic variation upon which natural selection can act. In a given organism, genetic variation can arise through point mutation, or by means of horizontal gene transfer. Should horizontal gene transfer have a role in spreading this adaptive mutation, A375L’s phylogenetic distribution would be random (independent of clade structures). To address this possibility, we performed a tBLASTn search on the conserved 16-residue motif AEAHHDGIETDSRLLT that carries the mutation (boldface underlined) and show that although this mutation exists elsewhere in nature, it is sparsely documented in GenBank, and always found in sporadic clades, which argues against its spread solely by horizontal gene transfer (Supplementary Figure S4). As this site evolved the same amino acid replacement in surface sequences repeatedly and independently, it is a clear candidate for validation by site-directed mutagenesis, which we are currently investigating; the functional implications are less clear as this site is located in a dimerisation domain found in pyridine nucleotide-disulphide oxidoreductase (Figure 4).
Evidence for selection on the mercuric reductase N-terminus. Location of site A375L that shows strongest signal of positive selection. (a) Three-dimensional model (PDB identifier: 4K7Z; chosen by SwissModel); secondary structure are indicated (green: β sheets; red: α helices), as well as position 375; the left and right structures are identical, but views differ by a rotation of 180 degrees; the region sequenced in this study is circled. (b) This position is shown in a subset of the protein sequence alignment for deepest (*DEP*), middle (*MID*) and surface (*SUR*) sequences; this position changes from an A to a T and to an L with time; AQSUR12 and AQSUR8 are among the three sequences where no positive selection could be detected (Supplementary Figure S3,Supplementary Information). (c) The identified Pfam domain is involved in the dimerisation of the MerA protein, here represented in its totality with its two Pfam domains, PF07992 and PF02852.
To further support this rapid evolutionary response to anthropogenic Hg emission, we analysed merA and rpoB data extracted from the three different depths using relaxed molecular clock models for different demographic scenarios (Drummond et al., 2006). Such demographic predictions are made possible by placing on the diversification process a prior distribution, that is function of the effective population size; the Bayesian modelling can then estimate these variables based on the shape of the estimated phylogenetic tree. Note that this modelling approach focuses on the effective size of populations of detoxifying prokaryotes (via the mer-operon sequence data), which differs from census data (as determined by quantitative PCR) as the effective size represents the number of individuals in an ‘ideal’ population that would produce the same amount of drift (see Supplementary Text for details on the modelling approach). Our demographic analyses were calibrated based on the absolute age of each depth (CE 2010, 1885 and <1800). Because the age of the deepest layer is only approximately known, we performed 12 separate analyses varying this calibration between 200 (CE 1810) and 2000 years ago (CE 10; Supplementary Text). Model fit was assessed by ranking each of these models according to their marginal likelihood  estimated by path sampling (Xie et al., 2011). These models fitted a flexible skyline demographic model that allowed the reconstruction of ancestral variation of effective population sizes, scaled by the generation length per year (Drummond et al., 2005); the scaled estimates can be understood as estimates directly proportional to the number of bacteria harbouring the merA gene. These skyline models, allowing for demographic fluctuations, were also ranked against models of constant population sizes. Our results demonstrate that (i) the skyline models fitted to the merA data outperform the constant population size models (Figure 2), (ii) that the age of the deepest layer is over 1100 years old (< CE 910;
estimated by path sampling (Xie et al., 2011). These models fitted a flexible skyline demographic model that allowed the reconstruction of ancestral variation of effective population sizes, scaled by the generation length per year (Drummond et al., 2005); the scaled estimates can be understood as estimates directly proportional to the number of bacteria harbouring the merA gene. These skyline models, allowing for demographic fluctuations, were also ranked against models of constant population sizes. Our results demonstrate that (i) the skyline models fitted to the merA data outperform the constant population size models (Figure 2), (ii) that the age of the deepest layer is over 1100 years old (< CE 910;  ; Supplementary Figure S5) and (iii) there was a significant increase in the scaled effective population size of the merA genes, ~220–170 years ago, coincident with the Industrial Revolution that took place 260–160 years ago (Figure 2, Supplementary Figure S6). While our housekeeping marker rpoB also shows evidence for a very old age at the deepest layer and a non-constant scaled effective population size (Supplementary Figure S7), the reconstructed demographics at this locus show a radically different pattern, with a very ancient and steady decrease in demography toward present times (Supplementary Figure S8). Therefore, our data support the assertion that the sedimentary record demographics of Hg-detoxifying apparatus reflect Hg speciation and levels at the time of deposition.
; Supplementary Figure S5) and (iii) there was a significant increase in the scaled effective population size of the merA genes, ~220–170 years ago, coincident with the Industrial Revolution that took place 260–160 years ago (Figure 2, Supplementary Figure S6). While our housekeeping marker rpoB also shows evidence for a very old age at the deepest layer and a non-constant scaled effective population size (Supplementary Figure S7), the reconstructed demographics at this locus show a radically different pattern, with a very ancient and steady decrease in demography toward present times (Supplementary Figure S8). Therefore, our data support the assertion that the sedimentary record demographics of Hg-detoxifying apparatus reflect Hg speciation and levels at the time of deposition.
A previous study of the MerA phylogeny suggested an early emergence of MerA among thermophilic bacteria thriving in hydrothermal environments with high Hg levels (Barkay et al., 2010), followed by continual gene recruitment improving efficiency of Hg detoxification (Boyd and Barkay, 2012). Until now, it has been very difficult, however, to relate these different steps in the evolution of the mer-operon over geological times to specific events in Earth’s history, except to earth oxygenation which may have supported diversification of mer-operon determinants (Barkay et al., 2010) and subsequent horizontal transfer to anaerobes or phototrophs. Indeed, only very few studies report mer-determinants in obligate anaerobes (Narita et al., 1999) or phototrophs (Marteyn et al., 2013) as they thrive in niches where Hg toxicity is thought to be decreased because of the absence of oxygen (Schaefer et al., 2002) or the presence of light (Gregoire and Poulain, 2014). Our findings suggest that although the mer-operon may have evolved for billions of years under geogenic Hg influence, anthropogenic Hg may now, at least partly, control its evolutionary path.
Our results support a rapid evolutionary response, over human time scales, of a gene involved in toxic metal resistance coinciding with changes in the delivery of this metal, at low levels, to remote ecosystems. We are currently working at retracing the evolutionary history of mercuric reductase genes using a combination of de novo gene synthesis and experimental evolution and testing whether (i) this signature solely reflects Hg speciation (that is, the chemical complexes that Hg forms with inorganic and organic ligands and how these complexes interact with cellular targets) and concentrations of Hg at the time of deposition, or (ii) also partially reflects microbial adaptation to their local environment once isolated from the surface.
These findings offer a novel way to investigate the fate and impact of globally distributed metals in the environment by assessing the response of microbes to changes in metal(loid)s availability and toxicity. We posit that what we observed for Hg could be true for any metal homeostatic or detoxification processes for which microbes have evolved regulation mechanisms offering potential for development of novel paleo-proxies tracking changes in the delivery of contaminants to ecosystems.
References
Abascal F, Zardoya R, Telford MJ . (2010). TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res 38: W7–W13.
Amos HM, Jacob DJ, Streets DG, Sunderland EM . (2013). Legacy impacts of all-time anthropogenic emissions on the global mercury cycle. Global Biogeochemical Cycles 27: 410–421.
Appleby P . (2001) Chronostratigraphic techniques in recent sediments. In: LAST WM, SMOL JP (eds) Tracking Environmental Changes in Lake Sediments: Physical and Chemical Techniques. Kluwer Academic Publishers: Dordrecht, The Netherlands.
Appleby PG, Oldfield F . (1978). The calculation of lead-210 dates assuming a constant rate of supply of unsupported 210Pb to the sediment. Catena 5: 1–8.
Barkay T, Kritee K, Boyd E, Geesey G . (2010). A thermophilic bacterial origin and subsequent constraints by redox, light and salinity on the evolution of the microbial mercuric reductase. Environ Microbiol 12: 2904–2917.
Barkay T, Miller SM, Summers AO . (2003). Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol Rev 27: 355–384.
Bertics VJ, Ziebis W . (2009). Biodiversity of benthic microbial communities in bioturbated coastal sediments is controlled by geochemical microniches. ISME J 3: 1269–1285.
Boere AC, Damste JSS, Rijpstra WIC, Volkman JK, Coolen MJL . (2011). Source-specific variability in post-depositional DNA preservation with potential implications for DNA based paleoecological records. Organic Geochemistry 42: 1216–1225.
Bordoli L, Schwede T . (2012). Automated protein structure modeling with SWISS-MODEL Workspace and the Protein Model Portal. Methods Mol Biol 857: 107–136.
Boyd ES, Barkay T . (2012). The mercury resistance operon: from an origin in a geothermal environment to an efficient detoxification machine. Front Microbiol 3: 349.
Brazeau ML, Poulain AJ, Paterson AM, Keller W, Sanei H, Blais JM . (2013). Recent changes in mercury deposition and primary productivity inferred from sediments of lakes from the Hudson Bay Lowlands, Ontario, Canada. Environ Pollut 173: 52–60.
Brigham ME, Sandheinrich MB, Gay DA, Maki RP, Krabbenhoft DP, Wiener JG . (2014). Lacustrine responses to decreasing wet mercury deposition rates-results from a case study in Northern Minnesota. Environ Sci Technol 48: 6115–6123.
Coolen MJL, Overmann J . (2007). 217 000-year-old DNA sequences of green sulfur bacteria in Mediterranean sapropels and their implications for the reconstruction of the paleoenvironment. Environ Microbiol 9: 238–249.
Coolen MJL, Talbot HM, Abbas BA, Ward C, Schouten S, Volkman JK et al. (2008). Sources for sedimentary bacteriohopanepolyols as revealed by 16S rDNA stratigraphy. Environ Microbiol 10: 1783–1803.
D'costa VM, King CE, Kalan L, Morar M, Sung WWL, Schwarz C et al. (2011). Antibiotic resistance is ancient. Nature 477: 457–461.
Delport W, Poon AF, Frost SD, Kosakovsky Pond SL . (2010). Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26: 2455–2457.
Drevnick PE, Canfield DE, Gorski PR, Shinneman ALC, Engstrom DR, Muir D et al. (2007). Deposition and cycling of sulfur controls mercury accumulation in isle Royale fish. Environ Sci Technol 41: 7266–7272.
Drummond AJ, Ho SYW, Phillips MJ, Rambaut A . (2006). Relaxed phylogenetics and dating with confidence. PLoS Biol 4: 699–710.
Drummond AJ, Rambaut A . (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 8.
Drummond AJ, Rambaut A, Shapiro B, Pybus OG . (2005). Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol 22: 1185–1192.
Edgar RC . (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797.
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R . (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200.
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR et al. (2014). Pfam: the protein families database. Nucleic Acids Res 42: D222–D230.
Glew JR . (1989). A new trigger mechanism for sediment samplers. Journal of Paleolimnology 2: 241–243.
Gough HL, Stahl DA . (2011). Microbial community structures in anoxic freshwater lake sediment along a metal contamination gradient. ISME J 5: 543–558.
Gregoire DS, Poulain AJ . (2014). A little bit of light goes a long way: the role of phototrophs on mercury cycling. Metallomics 6: 396–407.
Gunn J, Snucins E . (2010). Brook charr mortalities during extreme temperature events in Sutton River, Hudson Bay Lowlands, Canada. Hydrobiologia 650: 79–84.
Haile J, Holdaway R, Oliver K, Bunce M, Gilbert MTP, Nielsen R et al. (2007). Ancient DNA chronology within sediment deposits: Are paleobiological reconstructions possible and is DNA leaching a factor? Mol Biol Evol 24: 982–989.
Harris RC, Rudd JWM, Amyot M, Babiarz CL, Beaty KG, Blanchfield PJ et al. (2007). Whole-ecosystem study shows rapid fish-mercury response to changes in mercury deposition. Proc Natl Acad Sci USA 104: 16586–16591.
Hirsch PR, Mauchline TH, Clark IM . (2010). Culture-independent molecular techniques for soil microbial ecology. Soil Biology & Biochemistry 42: 878–887.
Inagaki F, Okada H, Tsapin AI, Nealson KH . (2005). Research paper: Microbial survival—The paleome: A sedimentary genetic record of past microbial communities. Astrobiology 5: 141–153.
Lauro FM, Demaere MZ, Yau S, Brown MV, Ng C, Wilkins D et al. (2011). An integrative study of a meromictic lake ecosystem in Antarctica. ISME J 5: 879–895.
Lejzerowicz F, Esling P, Majewski W, Szczucinski W, Decelle J, Obadia C et al. (2013). Ancient DNA complements microfossil record in deep-sea subsurface sediments. Biol Lett 9: 4.
Lyra C, Sinkko H, Rantanen M, Paulin L, Kotilainen A . (2013). Sediment bacterial communities reflect the history of a sea basin. PLoS One 8: 8.
Maidak BL, Cole JR, Lilbum TG, Parker CT, Saxman PR, Farris RJ et al. (2001). The Ribosomal Database Project (REP-II) at the Michigan State University in East Lansing, Michigan, release number 10. Nucleic Acids Res 29: 173.
Marteyn B, Sakr S, Farci S, Bedhomme M, Chardonnet S, Decottignies P et al. (2013). The synechocystis PCC6803 MerA-like enzyme operates in the reduction of both mercury and uranium under the control of the glutaredoxin 1 enzyme. J Bacteriol 195: 4138–4145.
Miller GE, Grant PM, Kishore R, Steinkruger FJ, Rowland FS, Guinn VP . (1972). Mercury concentrations in museum specimens of tuna and swordfish. Science 175: 1121–1122.
Morel FMM, Kraepiel AML, Amyot M . (1998). The chemical cycle and bioaccumulation of mercury. Annual Review of Ecology and Systematics 29: 543–566.
Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL . (2012). Detecting individual sites subject to episodic diversifying selection. PLoS Genet 8: e1002764.
Narita M, Huang C-C, Koizumi T, Endo G . (1999). Molecular analysis of merA gene possessed by anaerobic mercury-resistant bacteria isolated from sediment of Minamata Bay. Microbes and Environments 14: 77–84.
Osborn AM, Bruce KD, Strike P, Ritchie DA . (1997). Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol Rev 19: 239–262.
Parker J, Rambaut A, Pybus OG . (2008). Correlating viral phenotypes with phylogeny: Accounting for phylogenetic uncertainty. Infect Genet Evol 8: 239–246.
Parks JM, Johs A, Podar M, Bridou R, Hurt RA, Smith SD et al. (2013). The genetic basis for bacterial mercury methylation. Science 339: 1332–1335.
Paterson AM, Keller W, Rühland KM, Jones FC, Winter JG . (2014). An exploratory survey of water chemistry and plankton communities in lakes near the Sutton River, Hudson Bay Lowlands, Ontario, Canada. Arctic, Antarctic and Alpine Research 46: 121–138.
Posada D . (2008). jModelTest: Phylogenetic model averaging RID C-4502-2008. Mol Biol Evol 25: 1253–1256.
Poulain AJ, Ní Chadhain SM, Ariya PA, Amyot M, Garcia E, Campbell PGC et al. (2007). Potential for mercury reduction by microbes in the high Arctic. Appl Environ Microbiol 73: 2230–2238.
Price MN, Dehal PS, Arkin AP . (2009). FastTree: computing large minimum-evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641.
Rühland KM, Paterson AM, Keller W, Michelutti N, Smol JP . (2013). Global warming triggers the loss of a key Arctic refugium. Proc Biol Sci 280: 20131887.
Schaefer JK, Letowski J, Barkay T . (2002). mer-mediated resistance and Volatilization of Hg(II) under anaerobic conditions. Geomicrobiology Journal 19: 87–102.
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541.
Siciliano SD, O'driscoll NJ, Lean DRS . (2002). Microbial reduction and oxidation of mercury in freshwater lakes. Environ Sci Technol 36: 3064–3068.
Stoeva MK, Aris-Brosou S, Chetelat J, Hintelmann H, Pelletier P, Poulain AJ . (2014). Microbial community structure in lake and wetland sediments from a High Arctic Polar Desert revealed by targeted transcriptomics. PLoS One 9: 12.
Storey JD, Tibshirani R . (2003). Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445.
Takano Y, Tyler JJ, Kojima H, Yokoyama Y, Tanabe Y, Sato T et al. (2012). Holocene lake development and glacial-isostatic uplift at Lake Skallen and Lake Oyako, Lutzow-Holm Bay, East Antarctica: Based on biogeochemical facies and molecular signatures. Applied Geochemistry 27: 2546–2559.
Turpeinen R, Kairesalo T, Haggblom MM . (2004). Microbial community structure and activity in arsenic-, chromium- and copper-contaminated soils. FEMS Microbiol Ecol 47: 39–50.
UNEP. (2013). Global Mercury Assessment 2013: Sources, emissions, releases, and environmental transport (DTI/1636/GE).
Xie WG, Lewis PO, Fan Y, Kuo L, Chen MH . (2011). Improving marginal likelihood estimation for Bayesian phylogenetic model selection. Syst Biol 60: 150–160.
Zhou JZ, Bruns MA, Tiedje JM . (1996). DNA recovery from soils of diverse composition. Appl Environ Microbiol 62: 316–322.
Acknowledgements
This work was funded by NSERC Discovery grants to AJP, JMB and SAB, the Ontario Ministry of the Environment and Climate Change through the Climate Change and Multiple Stressor Research program at Laurentian University, and the Ontario Graduate Scholarship Program. We thank Tamar Barkay, Lars Dietrich, Dianne Newman, Alexa Price-Whelan, John Smol and Paula Welander for comments on previous versions of this manuscript. We thank Albert and Gilbert Chookomolin for their assistance in the field. We thank Dr Emmanuel Yumvihoze, Philip Pelletier and Maggie Stoeva for their help with lab analyses and Linda Kimpe for 210Pb analyses.
Author Contributions
AJP initiated the study; AJP, JMB and SAB designed the experiments and AJP, MB, AP and WK carried them out; AJP and SAB contributed equally to this work, carried out data analysis and wrote the manuscript with inputs from all co-authors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Poulain, A., Aris-Brosou, S., Blais, J. et al. Microbial DNA records historical delivery of anthropogenic mercury. ISME J 9, 2541–2550 (2015). https://doi.org/10.1038/ismej.2015.86
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2015.86
This article is cited by
-
Mining-impacted rice paddies select for Archaeal methylators and reveal a putative (Archaeal) regulator of mercury methylation
ISME Communications (2023)
-
Comparative genomics revealed that Vibrio furnissii and Vibrio fluvialis have mutations in genes related to T6SS1 and T6SS2
Archives of Microbiology (2023)
-
Sediment-associated microbial community profiling: sample pre-processing through sequential membrane filtration for 16S rRNA amplicon sequencing
BMC Microbiology (2022)
-
Swift evolutionary response of microbes to a rise in anthropogenic mercury in the Northern Hemisphere
The ISME Journal (2020)
-
Cellular toxicity pathways of inorganic and methyl mercury in the green microalga Chlamydomonas reinhardtii
Scientific Reports (2017)