Abstract
The goal of phytoremediation is to use plants to immobilize, extract or degrade organic and inorganic pollutants. In the case of organic contaminants, plants essentially act indirectly through the stimulation of rhizosphere microorganisms. A detailed understanding of the effect plants have on the activities of rhizosphere microorganisms could help optimize phytoremediation systems and enhance their use. In this study, willows were planted in contaminated and non-contaminated soils in a greenhouse, and the active microbial communities and the expression of functional genes in the rhizosphere and bulk soil were compared. Ion Torrent sequencing of 16S rRNA and Illumina sequencing of mRNA were performed. Genes related to carbon and amino-acid uptake and utilization were upregulated in the willow rhizosphere, providing indirect evidence of the compositional content of the root exudates. Related to this increased nutrient input, several microbial taxa showed a significant increase in activity in the rhizosphere. The extent of the rhizosphere stimulation varied markedly with soil contamination levels. The combined selective pressure of contaminants and rhizosphere resulted in higher expression of genes related to competition (antibiotic resistance and biofilm formation) in the contaminated rhizosphere. Genes related to hydrocarbon degradation were generally more expressed in contaminated soils, but the exact complement of genes induced was different for bulk and rhizosphere soils. Together, these results provide an unprecedented view of microbial gene expression in the plant rhizosphere during phytoremediation.
Similar content being viewed by others
Introduction
The rhizosphere comprises the surface of the roots and the surrounding soil area where plant root exudates sustain a high microbial activity and high microbial density (Smalla et al., 2001; Kowalchuk et al., 2002). However, bacterial diversity in the rhizosphere is generally lower than in the bulk soil (Marilley and Aragno, 1999), and microbial community composition is very different (Smalla et al., 2001; Kowalchuk et al., 2002; Griffiths et al., 2006; Kielak et al., 2008), suggesting a strongly selective environment. This selection pressure results from the exudation of specialized antimicrobials and signaling molecules (for example, flavonoids, salicylic acid and phytoalexins), carbon (for example, organic acids and aromatic compounds) and nitrogen (for example, amino acids) compounds. The rhizosphere is thereby selectively enriched in microorganisms that are adapted to highly competitive environments and to the utilization of specific plant compounds (Berg et al., 2002; Gomes et al., 2003; Berg et al., 2005; Haichar et al., 2008). These compounds are not only exuded for the benefit of microbes, but more often they profit the plant itself. For instance, organic acids such as malate, citrate and oxalate are often present in the rhizosphere, and in addition to being a carbon source for many microbes, they are involved in many plant processes such as metal detoxification, nutrient acquisition and alleviation of stress (Jones, 1998). Interactions in the rhizosphere have evolved over millions of years and can be seen as a way for plants to reach a minimal stress level by, among others, deterring pathogens, increasing their nitrogen and phosphorus uptake and detoxifying the environment. Plants confronted with stressful environments normally respond by increasing root exudation (Jones et al., 2004; Qin et al., 2007; Naik et al., 2009), which leads to increased microbial biomass in the rhizosphere (Esperschutz et al., 2009).
One biotechnological application of the ‘rhizosphere effect’ is phytoremediation. The goal of phytoremediation is to remove pollutants from the environment or render them harmless by using plants to stabilize, filter, volatilize, extract or degrade organic and inorganic pollutants (Salt et al., 1998; Pilon-Smits, 2005). For the degradation of organic contaminants, plants essentially act indirectly through the specific stimulation of rhizosphere and endophytic microorganisms (Barac et al., 2004; Kuiper et al., 2004; Taghavi et al., 2005). Phytoremediation takes advantage of the long-evolved intimate relationships between plant and microbes, of the stimulating effect of plants on microbes and of the natural hardiness and competitiveness of rhizosphere microbes to remediate contaminated soils or to mobilize inorganic contaminants and favor their accumulation in plant tissues. It is one of the least expensive and most environmentally friendly remediation techniques, being on average tenfold less expensive than traditional excavation techniques (‘dig and dump’) (Glass, 1999) and causing very little disruption to the environment. However, phytoremediation often proceeds slowly, as the stimulation of degrading microbes is mainly restricted to the immediate zone of influence of the roots (Pilon-Smits, 2005). For these reasons, other approaches are often preferred and phytoremediation is restricted to niche markets where time is not an issue and the contamination is moderate and superficial. To avoid this limitation, fast-growing trees that rapidly develop deep-root systems and produce large biomass, such as poplars and willows, were suggested (Schnoor et al., 1995). Willow trees have several key advantages for phytoremediation when compared with other plants: they are genetically very diverse (400 species and over 200 hybrids, Newsholme, 2003), some species can be harvested frequently by coppicing, they are pioneer plants that have invasive growth strategies and very effective nutrient uptake systems, they grow fast and have high evapotranspiration rates and high productivity (Pulford and Watson, 2003). Although several studies have assessed the microbial communities associated with willows growing in contaminated soils (Leigh et al., 2006; de Carcer et al., 2007a, 2007b; Kuffner et al., 2008; Hrynkiewicz et al., 2009; Zimmer et al., 2009; Weyens et al., 2013; Bell et al., 2014a), the details of willow interactions with microbes are still not well understood.
Clearly, plant–microbe interactions are at the center of the phytoremediation process of organic contaminants and have a role in the mobilization of inorganic contaminants, but which rhizosphere genes and organisms are involved and how these complex interactions are affected by contaminant type and concentration have not been elucidated. This knowledge is crucial to fully optimize the degradation processes that occur in the rhizosphere. In this study, our main objective was to understand what microorganisms are activated and what microbial genes are upregulated in the rhizosphere of willow growing in contaminated and non-contaminated soils as compared with bulk soil. We harvested the rhizosphere of willows planted in pots containing contaminated and non-contaminated soil, as well as soil from non-planted pots. Gene expression profiles were contrasted using metatranscriptomic analyses based on Illumina sequencing, and microbial communities were compared based on Ion Torrent sequencing of 16S rRNA. Our results indicate major shifts in the potential for carbon and amino-acid uptake, and utilization, nutrient cycling and hydrocarbon degradation in the rhizosphere of willows, with a much stronger stimulation in contaminated soils. To our knowledge, this is the first functional metatranscriptomic profiling of microbial activities in the rhizosphere, providing an unprecedented level of detail on plant–microbial interactions.
Materials and methods
For more details, see the Supplementary Material and Methods.
Greenhouse experiment
Contaminated and non-contaminated soils were collected at a former petrochemical plant site at Varennes, QC, Canada (geographic coordinates: contaminated: 45.699145 and −73.430997, and non-contaminated: 45.700788 and −73.430302). Various petrochemical activities had been carried out on this site, starting in 1953 to the shutdown of the plant in 2008. The soils have been contaminated for decades by mixed petrochemical residues (see Supplementary Table S1 for detailed soil analyses). After sampling, soils were mixed thoroughly and distributed in 20 l pots. Willow cuttings (Salix purpurea cultivar Fish Creek) were first grown for 8 weeks in sterile potting media and then transferred to the pots containing Varennes soil on 4 October 2011. At the time of the transfer, the longest stem that grew from the cuttings was ∼20 cm long and the potting media was densely colonized by roots. Half of the pots remained unplanted and each treatment was replicated six times. The experimental design was a split-plot with soil contamination randomized first and planted or unplanted randomized in the subplots. The plants were grown in a greenhouse at the Institut de recherche en biologie végétale, Montreal, under natural daylight supplemented with high-pressure sodium-vapor lamps, and temperatures of 20 °C in day and 18 °C at night. The moisture in every pot was maintained near-field capacity by frequent watering, and saucers were used under pots to prevent leaching of contaminants.
Soil sampling
For molecular analyses, pots were sampled at the end of the experiment (26 April 2012, ∼6 months after planting). For rhizosphere soil, plants were completely recovered from the pots and shaken vigorously to remove excess soil. Soil still adhering to the roots at this stage was considered as rhizosphere soil. Soil from unplanted pots was taken at a depth of ∼5 cm. At least five different soil subsamples were collected from each pot and homogenized in 50 ml Falcon tubes and immediately flash frozen in liquid nitrogen. In total, the entire process for one pot never took more than a few minutes. Tubes were transported from the greenhouse to the lab under dry ice and kept frozen at −80 °C until the nucleic acid was extracted. For chemical analyses, pots were sampled at the beginning and at the end of the experiment. From each pot, soil was collected from at least five different zones and homogenized in an amber glass container. Soil samples were sent to Maxxam Analytics (Montreal, QC, Canada), where soil was analyzed for C10–C50 hydrocarbons (sum of all aliphatic hydrocarbon compounds with chain lengths from C10–C50) and polycyclic aromatic hydrocarbons (PAHs) according to standard protocols. The percentage degradation was calculated separately for each of the pots.
Nucleic acid extraction
Approximately 2 g of frozen soil was weighed and extracted using MoBio RNA PowerSoil total RNA isolation kit with the RNA PowerSoil DNA elution accessory kit (MoBio, Carlsbad, CA, USA). RNA extracts were treated with Ambion TURBO DNAse (Life Technologies, Burlington, ON, Canada) and the absence of DNA was confirmed by 16S rRNA gene universal PCR.
Ion Torrent 16S rRNA sequencing
Reverse-transcriptase (RT)-PCR of the partial 16S rRNA was performed using the universal primers F343 and R533 containing the 10-bp multiplex identifiers and adaptor sequences for Ion Torrent sequencing described previously (Yergeau et al., 2012; Bell et al., 2013b). The sequencing of the pooled library was done using the Ion Torrent Personal Genome Machine system (Life Technologies). Sequences were binned and filtered using a custom-made Perl script. Taxonomic identities were assigned to sequences using the ‘multiclassifier’ (http://pyro.cme.msu.edu/). Weighted-normalized Unifrac distances between each sample pair were calculated using the FastUnifrac website (Hamady et al., 2010) based on the GreenGene core data set.
Illumina mRNA sequencing
rRNA was subtracted following the protocol described by Stewart et al. (2010). Total rRNA-subtracted RNA was reverse-transcribed using the SuperScript III kit (Invitrogen). Illumina libraries were prepared following the protocol of Meyer and Kircher (2010), with indices 1–24 pooled together and sent for eight lanes of Illumina HiSeq 2000 paired-end 2 × 101 bp sequencing at McGill University and Génome Québec Innovation Center, Montréal. Data from the different lanes were pooled together and the resulting 48 files were filtered in pairs using a custom-made Perl script. The resulting high-quality sequences were submitted to MG-RAST 3.0 (Meyer et al., 2008) for automated annotation.
Data analysis
All statistical analyses were carried out in R (v 2.13.2, The R Foundation for Statistical Computing). Normal distribution and variance homogeneity of the data were tested using the ‘shapiro.test’ and ‘bartlett.test’ functions, respectively. If the data were not normally distributed or did not show homogeneous variance, they were log transformed before analysis of variance (ANOVA) analyses. ANOVA were carried out using the ‘aov’ function, whereas t-test were performed using the ‘t.test’ function. Multivariate tests of hypothesis were carried out using Permanova with the ‘adonis’ function of the ‘vegan’ package. The Unifrac matrix was used for principal coordinate analyses (PCoA) that were carried out using the ‘pcoa’ function of the ‘ape’ package.
Results
Soil physico-chemical characteristics and plant biomass
The non-contaminated soils contained no PAH and no C10–C50 hydrocarbons at the beginning of the experiment (Table 1; see Supplementary Table S1 for a description of the contaminants). There was only one exception in the to-be planted pots, where C10–C50 hydrocarbons were detected at a level of 110 mg kg−1 (Supplementary Table S1). At the end of the experiment, all the soils in the non-contaminated pots had C10–C50 and PAH concentrations below the detection limit. For the contaminated soils, no significant differences at P<0.05 (t-test) were observed between the planted and unplanted pots at the beginning of the experiment or at the end for both C10–C50 and PAHs (Table 1, Supplementary Table S1). The concentrations of C10–C50 and PAHs were more variable at the beginning of the experiment (C10–C50: 480–4200 mg kg−1; PAH: 15.6–186.4 mg kg−1) than at the end of the experiment (C10–C50: 270–660 mg kg−1; PAH: 3.0–19.3 mg kg−1). There was a large decrease in hydrocarbon concentrations in all contaminated soils during the incubation period, but there was no significant difference between planted and unplanted pots at P<0.05 (t-test), with degradation ranging from 0% to 91% for C10–C50 and from 65% to 93% for PAH.
At the final time of sampling (April 2012), contaminated and non-contaminated soils from the unplanted pots and from the rhizosphere of willows were sampled for chemical analyses. Water-extractable PO4 was not detected in any of the samples, whereas the concentration of water-extractable K was significantly higher in contaminated soil independent of the soil compartment (379 μM kg−1 vs 261 μM kg−1), and the concentration of water-extractable NO3 was significantly lower in the rhizosphere of willows independent of contamination levels (59.5 μM kg−1 vs 454.9 μM kg−1) (B Cloutier-Hurteau, M-C Turmel and F Courchesne, personal communication). For willow biomass, there was no significant difference between willows growing in contaminated or non-contaminated soils for the shoots (average: 36.0 g), the roots (average: 6.9 g) and total plant biomass (average: 50.3 g), but the willows growing in contaminated soils had significantly higher leaf biomass (8.4 g vs 6.3 g) (B Cloutier-Hurteau, M-C Turmel and F Courchesne, personal communication).
Carbon and amino-acid utilization
The expression of several gene categories (MG-RAST ‘level 3’) related to carbon uptake and utilization was significantly influenced by the presence of plants and soil contamination levels (Figure 1a). Most gene categories were more actively expressed in the rhizosphere of contaminated soils, including all gene categories related to organic acids (Figure 1a). The non-planted soils clustered together and shared similar expression levels of several gene categories, but the rhizosphere soils showed very dissimilar patterns of expression and did not cluster together (Figure 1a). Permanova tests for the effect of the different treatments on all the gene categories selected yielded significant results for contamination level (F=4.75, P=0.0010) and willow presence (F=7.43, P=0.0001), but not for the interaction between contamination and willow. Based on the F-ratios, the strongest effect was seen when comparing non-planted vs rhizosphere soils. The expression of the phosphoenolpyruvate carboxykinase (pckA) gene, which encodes the enzyme that catalyses the key rate-limiting step in gluconeogenesis, was significantly induced by willow presence (F=7.40, P=0.015), but not by contamination level (Figure 2a). Similarly, the expression of carbohydrate uptake ABC transporter (CUT1 family), which is usually involved in the import of oligosaccharides and their derivatives, was significantly enhanced in the rhizosphere of willows (F=7.19, P=0.015), but not affected by contamination levels (Figure 2a).
Carbohydrate (a) and amino-acid (b) uptake and utilization potential patterns for contaminated (C) or non-contaminated (NC) rhizosphere (P) or bulk (NP) soil. The heatmaps are drawn from centered-scaled normalized mRNA abundance data averaged for each treatment (N=6); the darker the cell, the higher the mRNA abundance. The MG-RAST ‘hierarchical classification’ functionality was used with the M5NR database and, from the level 1 ‘carbohydrates’ and ‘amino acids and derivative’ gene categories, all level 3 gene categories that contained the keywords ‘utilization’, ‘degradation’, ‘catabolism’, ‘uptake’ were selected.
Normalized mRNA abundance for (a) phosphoenolpyruvate carboxykinase (pckA) and carbohydrate uptake ABC transporter (CUT1) and (b) quorum sensing and biofilm formation gene category and antibiotic resistance genes for contaminated (C) or non-contaminated (NC) rhizosphere (P) or bulk (NP) soil. For (a), the MG-RAST ‘all annotation’ functionality with the GenBank database was used to search for the genes of interest. For (b), the MG-RAST ‘hierarchical classification’ functionality was used with the M5NR database and the ‘Quorum sensing and biofilm formation’ level 2 gene category or the antibiotic resistance-related gene categories from the ‘Resistance to antibiotics and toxic compounds’ level 2 gene category were summed. Values are normalized abundances, and error bars are s.d. (N=6). **P<0.01; ***P<0.001; n.s: not significant.
Most gene categories (MG-RAST ‘level 3’) related to amino-acid utilization and degradation were more expressed in the rhizosphere of willows planted in contaminated soils (Figure 1b). This was also true for the degradation of aromatic amino acids such as tryptophan and histidine (Figure 1b). In the case of histidine degradation, this gene category was also highly expressed in the rhizosphere of willows planted in non-contaminated soils (Figure 1b). The non-contaminated soils clustered together, whereas the contaminated willow rhizosphere soil was a clear outlier (Figure 1b). Permanova tests for the effect of treatments on the overall expression pattern of amino-acid utilization gene categories resulted in a significant interaction term (F=5.28, P=0.002), suggesting a differential effect of willows in contaminated and non-contaminated soils. Contamination level and willow presence also had a significant influence on gene expression patterns (F=4.69, P= 0.0034 and F=6.63, P=0.00030, respectively). Based on the F-ratios, the strongest effect was seen when comparing non-planted vs rhizosphere soils.
Competition and cooperation
There was a significant effect of contamination level (F=16.15, P=0.00067) and willow presence (F=29.33, P=0.000027), but not of the interaction term on the expression of genes related to antibiotic resistance. Antibiotic resistance genes were more expressed in the rhizosphere of willow across both contamination levels (Figure 2b). Similarly, antibiotic resistance genes were significantly more expressed in contaminated soils, regardless of willow presence. These two trends resulted in a higher expression of antibiotic resistance genes in the rhizosphere of willows planted in contaminated soils as compared with all other treatments (Figure 2b). The expression of quorum sensing and biofilm formation genes was significantly affected by contamination level (F=26.14, P=0.000053), willow presence (F=23.47, P=0.000098) and the interaction term (F=8.40, P=0.0089).
Active microbial community composition
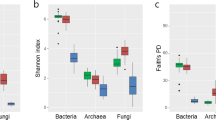
Two methods were used to determine active microbial community composition: taxonomic classification of all sequenced mRNA in MG-RAST and 16S rRNA sequencing. The first method, with the normalization used in this study, gives the activity (total abundance of mRNA) for different taxa, whereas the second method gives an overview of the active community composition with relative activity values (percentage of all ribosomes) for different taxa. There was a significant effect of contamination level (F=27.22, P= 0.000042) and the interaction term (F=17.14, P=0.00051) on the total abundance of mRNA related to Bacteria (bacterial activity), whereas the effect of willow was marginally significant (F=3.36, P=0.082) (Figure 3a). This was confirmed by RT-quantitative PCR based on bacterial 16S rRNA, which identified the contaminated willow rhizosphere soil as having significantly higher bacterial activity. In contrast, fungi were significantly more active in the rhizosphere of willows (F=43.22, P= 0.0000021) as compared with bulk soil, and the interaction term was significant (F=7.36, P=0.013), whereas contamination level had no significant effect (Figure 3a). This different rhizosphere effect on bacterial and fungal activity was also visible at the phylum/class level, where most fungal phyla (Basidiomycota, Ascomycota and Glomeromycota) were more active in the rhizosphere of willows planted in non-contaminated soils (Figure 3b). Actinobacteria and Firmicutes were also more active in the rhizosphere of willows planted in non-contaminated soils. In contrast, several of the most abundant bacterial phyla/classes (Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria and Acidobacteria) were more active in the rhizosphere of willows planted in contaminated soils (Figure 3b). Most phyla/classes showed low activity in the bulk soil, and contaminated and non-contaminated bulk soils clustered together (Figure 3b). Euryarchaeota were more active in the contaminated bulk soil, whereas Crenarchaeota and Thaumarchaeota were more active in the rhizosphere of willows planted in non-contaminated soil (Figure 3b). When testing the effects of the treatments on the activities at the phylum/class level using Permanova, the interaction term was significant (F=9.20, P=0.0001) as well as both single factors (contamination level: F=4.61, P=0.0044, willow presence: F=13.39, P=0.0001), with a dominance of the willow effect (higher F-ratio).
Microbial activities per taxa, based on total mRNA. (a) Normalized abundance of bacterial and fungal-related mRNA. Values are normalized abundances, and error bars are s.d. (N=6). *P<0.05; ***P<0.001; n.s.: not significant. (b) Heatmap for different phyla/classes drawn from centered-scaled normalized mRNA abundance data averaged for each treatment (N=6); the darker the cell, the higher the mRNA abundance.
The significant differences observed at the phylum/class level persisted at lower taxonomic levels. At the genus level, the interaction term (F=9.60, P=0.0001) and both single factors (contamination: F=4.74, P=0.0021; willow presence: F=6.96, P=0.0003) were significant in Permanova tests. The effect of willow presence was then tested on each genus, separately for contaminated and non-contaminated soils (Supplementary Table S2). The activities of several genera were significantly affected by willows both in contaminated and non-contaminated samples. However, more genera were significantly affected by willows in the non-contaminated soils (78 less active with willows and 1963 more active with willows, total 2041) than in the contaminated soils (8 less active with willows and 110 more active with willows, total 118). Among these, 42 genera were significantly affected by willows at both contamination levels, and the 24 bacterial and fungal genera among those are listed in Supplementary Table S2. Among the genera consistently activated by willows at both contamination levels, some were previously reported as having interesting characteristics for phytoremediation, for instance, Methylibium petroleiphilum is involved in aromatic hydrocarbon and methyl tert-butyl ether degradation (Nakatsu et al., 2006), Mesorhizobium is a well-known genus involved in nitrogen fixation, some species are resistant to metals (Vidal et al., 2009; Huang et al., 2010) and can degrade acetonitrile (Feng and Lee, 2009), some Variovorax species display plant growth promoting activities and are able to degrade various contaminants (Han et al., 2011) and Parvibaculum species are able to degrade linear carbon chains, including alkanes (Schleheck et al., 2004; Rosario-Passapera et al., 2012). Some other genera showed contrasting responses to willows when exposed to different concentrations of contaminants (Supplementary Table S2). Most of the genera that were positively affected by willows in contaminated soils and negatively affected in non-contaminated soils belonged to the Alphaproteobacteria or the Gammaproteobacteria, whereas the two genera negatively affected by willows in contaminated soils, but positively affected in non-contaminated soil belonged to the Blastocladiomycota (Supplementary Table S2).
The active community composition based on 16S rRNA gene sequencing revealed that contamination level was the main influencing factor (Figure 4a). However, within this dominant effect of contamination levels, willow presence did have some effect on bacterial community composition. At lower taxonomic levels (Unifrac distances), willow presence only had a significant effect on microbial communities in the contaminated soils (Permanova: non-contaminated soils: F=1.29, P=0.14; and contaminated soils: F=1.95, P=0.016). This effect can also be visualized in the principal coordinate ordinations (Figure 4b). In contrast, active species richness (number of operational taxonomical units at 97% similarity) was not affected by willow presence or contamination levels.
Active microbial community composition based on 16S rRNA gene sequencing for contaminated (C) or non-contaminated (NC) rhizosphere (P) or bulk (NP) soil. (a) Community composition at the phylum/class level averaged for each treatment (N=6). (b) PCoA based on Unifrac distances between samples for non-contaminated (NC) and contaminated (C) soils.
Nitrogen cycle genes
Nitrogen cycling is one of the key soil functions in the context of soil bioremediation, where nutrients are often limiting due to an unbalanced C:N ratio. Willow significantly increased the expression of ammonia monooxygenase genes, as willow presence was the only significant factor in ANOVA tests (F=5.68, P=0.027) (Figure 5a). Willows presence appeared to have a stronger effect on ammonia monooxygenase expression in non-contaminated soils (Figure 5a). Willow had a significant influence on the expression of several other genes, with an increased expression of genes related to nitrogen fixation (nitrogenase) and nitrate reduction in the rhizosphere, and higher expression of several genes related to denitrification in the bulk soils (Figure 5b). The overall effect of the treatments on the different nitrogen cycle genes was tested by Permanova, which highlighted significant effects of willow presence (F=2.39, P=0.034), contamination levels (F=2.75, P=0.016) and the interaction term (F=2.46, P=0.027).
Activities related to the nitrogen cycle for contaminated (C) or non-contaminated (NC) rhizosphere (P) or bulk (NP) soil. (a) Normalized abundance for ammonia monooxygenase (amoA)-related mRNA. The MG-RAST ‘all annotation’ functionality with the GenBank database was used to search for the genes of interest. Values are normalized abundances and error bars are s.d. (N=6). *P<0.05; n.s.: not significant. (b) Heatmap for different gene categories related to the N-cycle drawn from centered-scaled normalized mRNA abundance data averaged for each treatment; the darker the cell, the higher the mRNA abundance. The MG-RAST ‘hierarchical classification’ functionality was used with the M5NR database and from the level 1 ‘Nitrogen metabolism’ gene category functions that had key roles in the N-cycle, were selected.
Hydrocarbon degradation genes
The expression of alkane hydroxylase genes was significantly affected by contamination level (F=23.50, P=0.000098), willow presence (F=10.52, P=0.0041) and the interaction term (F=5.73, P=0.027), with a dominating effect of contamination (higher F-ratio) (Figure 6a). Alkane hydroxylases genes were more expressed in the rhizosphere of willows planted in contaminated soils than in any other treatment (Figure 6a). For the expression of genes related to the degradation of aromatic compounds, Permanova tests revealed a strong effect of contamination (F=7.76, P=0.0001) and of willow presence (F=3.90, P=0.0056), whereas the interaction term was significant but with a weaker effect (F=2.63, P=0.030). Within contaminated soils, willows induced the expression of several aromatic degradation gene categories and pathways, whereas some others were repressed by the presence of willows and others were highly expressed across contaminated soils (Figure 6b). With a few exceptions, the vast majority of aromatic hydrocarbon degradation gene categories showed relatively low expression levels in the non-contaminated soils (Figure 6b).
Activities related to hydrocarbon degradation for contaminated (C) or non-contaminated (NC) rhizosphere (P) or bulk (NP) soil. (a) Normalized abundance for alkane hydroxylase-related mRNA. The MG-RAST ‘all annotation’ functionality with the GenBank database was used to search for the genes of interest. Values are normalized abundances, and error bars are s.d. (N=6). *P<0.05; **P<0.01; ***P<0.001. (b) Heatmap for different gene categories related to the metabolism of aromatic compounds drawn from centered-scaled normalized mRNA abundance data averaged for each treatment (N=6); the darker the cell, the higher the mRNA abundance. The MG-RAST ‘hierarchical classification’ functionality was used with the M5NR database and from the level 1 ‘Metabolism of aromatic compounds’ all level 3 gene were used.
Discussion
Several studies have addressed the transcriptomics of the rhizosphere (Diehn and Relman, 2001; Mark et al., 2005; Matilla et al., 2007; Ramachandran et al., 2011), but always using a controlled approach with a single bacterium, one or a few plants in sterile substrates or purified root exudates. In this study, we used a metatranscriptomic approach and assessed the whole microbial community associated with willow roots growing in non-sterile soil from a contaminated site. This extremely powerful approach has often been proposed (Kowalchuk et al., 2010; Schenk et al., 2012; Turner and Poole, 2013) and was recently applied to examine taxonomic changes in the rhizosphere (Turner et al., 2013) but, to our knowledge, this is the first time it has been used to assess the functions, activities and the interactions in the rhizosphere at the community level. The results presented here give, with an unprecedented level of detail, a portrait of the microbial activities in the rhizosphere of willows as compared with bulk soil, and link these shifts to the potential for phytoremediation of organic contaminants by willows.
Metatranscriptomics of the rhizosphere
Although the composition of the active microbial community (based on 16S rRNA genes; Figure 4) did not vary much with the introduction of willows, being more strongly affected by the contamination levels, the community members drastically changed their activities (based on mRNA; Figure 3). Microbes in the rhizosphere of willow had significantly different expression patterns for genes involved in carbon and amino-acid uptake and utilization, strong, indirect evidence for the use of compounds in root exudates. The overall increase in the expression of phosphoenolpyruvate carboxykinase (involved in the rate-controlling step of glucose synthesis; repressed by sugars) and CUT1 transport gene (involved in the transport of oligosaccharides) expression in the rhizosphere and the high expression of organic acid uptake and utilization genes in the contaminated rhizosphere suggest that simple sugars are not the main bacterial carbon source in the rhizosphere of willows. Organic acids were previously reported as the main source of carbon in the rhizosphere of many plants, including poplar (Jones, 1998; Lugtenberg et al., 2001; Naik et al., 2009; Ramachandran et al., 2011).
The effect of willow presence varied depending on the contamination level of the soil. For instance, bacteria appeared to be generally stimulated in contaminated soils along with many functions, whereas fungi were more clearly stimulated in non-contaminated soils. Similarly, the rhizosphere effect of birch on soil microbial communities was reported to depend on PAH concentration (Sipilä et al., 2008). One possible explanation for this result is that plants recruit the rhizosphere microbial communities from the adjacent bulk soil (de Ridder-Duine et al., 2005; Dennis et al., 2010; Lundberg et al., 2012), and that contaminated and non-contaminated soils harbored different starting microbial communities (Figure 4). Along with specialized carbon compounds, plants also exude several antimicrobial compounds that create a strongly selective environment in the rhizosphere (Bais et al., 2006). Several studies reported decreased microbial diversity in the rhizosphere (Marilley and Aragno, 1999; Kowalchuk et al., 2002), supporting the concept of a more selective environment. In our case, the presence of contaminants in the soils is imposing a further selection pressure on microorganisms. Under this double-selection pressure, only microorganisms that can use specialized carbon sources, can cope with the presence of contaminants and are highly competitive will be significantly activated by willows. Our results showed that more microbial genera were activated by willows in non-contaminated soils than in contaminated soils. We also observed significant increases in the expression of antibiotic and toxic compound resistance genes in the rhizosphere, with stronger increases in contaminated soils, alluding to increased microbial competition or a less hospitable rhizosphere environment. Plants exude antimicrobial compounds (for example, phytoalexins, salicylic acid and flavonoids) that can lead to increased expression of antibiotic resistance genes. Salicylic acid was reported to induce the expression of antibiotic resistance genes in Escherichia coli (Cohen et al., 1993), whereas multidrug efflux pump genes of Rhizobium etli were induced by bean root flavonoids and were needed for resistance to phytoalexins, flavonoids and salicylic acid, and effective colonization of the rhizosphere (Gonzalez-Pasayo and Martinez-Romero, 2000). Mutation in multidrug efflux pumps reduced the fitness of Pseudomonas putida in the rhizosphere of corn (Matilla et al., 2007). Increased selection pressure in the contaminated rhizosphere soil also potentially led to an increased expression of genes related to cooperation and communication (quorum sensing and biofilm formation category). Biofilm formation is an essential feature of rhizosphere colonization (Lugtenberg et al., 2001; Ramey et al., 2004) and bacteria living in biofilms have increased resistance to environmental stresses (Costerton et al., 1999; Hogan and Kolter, 2002). Consequently, genes related to biofilm formation were reported to be over-expressed during rhizosphere colonization, and their inactivation led to reduced fitness (Matilla et al., 2007; Ramachandran et al., 2011; Fan et al., 2012). Some of the genes included in the quorum sensing and biofilm formation category (for example, N-acyl homoserine lactone synthetase) are also involved in plant–microbe communication, as N-acyl homoserine lactone will induce plants to release root exudates (Mathesius et al., 2003).
In the present study, we observed an extremely pronounced induction of carbon uptake and utilization genes in the rhizosphere of willows growing in contaminated soils, indirectly suggesting an increased exudation, which could explain the different rhizosphere effects in contaminated and non-contaminated soils. Poplars root exudation of C was stimulated up to 100-fold by increasing Al, Cu and Zn concentrations (Qin et al., 2007; Naik et al., 2009). These increases in the exudation are mechanisms by which plants reduce the toxicity of trace elements in soil, as organic acids can effectively bind many compounds. Stressed beech trees were also shown to have a larger microbial biomass in their rhizosphere (Esperschutz et al., 2009). Our contaminated soils did contain a range of trace elements, but at relatively low concentrations, in the same range as the non-contaminated soils (B Cloutier-Hurteau, M-C Turmel and F Courchesne, personal communication), which is unlikely to have caused significantly higher stress to the plants in the contaminated soils. We also did not observe any clear indication of an increasingly stressful environment for the plants growing in contaminated soils. In fact, plant biomass was not reduced in contaminated soils and leaf biomass even increased when willows were planted in contaminated soils. As suggested by Henry et al. (2007), one way to improve phytoremediation would be by increasing plant stress levels through various manipulations (for example, soil nutrient and water content), in order to increase stimulation of the microbes in the rhizosphere.
The concentration of nitrate was lower in the rhizosphere of willows planted in both contaminated and non-contaminated soil. The chemical data matched well with the observed patterns in the expression of genes related to the nitrogen cycle: high nitrate concentration is conducive to the expression of denitrification genes (Barnard et al., 2005) (several of these genes were more expressed in the bulk soil) and inhibits nitrogen fixation (Streeter and Wong, 1988) (these genes were highly expressed in the rhizosphere). The lower concentration of nitrate might be because of the increased microbial activity observed in the rhizosphere, especially in contaminated soils, where bacteria were significantly more active and nitrate at its lowest concentration, being below the detection limit for two of the three replicates analyzed. Nitrate reduction was shown to be stimulated by root exudates (Henry et al., 2008), and, in this study, several nitrate reductase genes were more expressed in the rhizosphere of willow, offering another potential explanation for the lower nitrate concentration observed in the rhizosphere. Taken together, these data suggest that the stimulation of the rhizosphere hydrocarbon-degrading microorganisms by willows is not because of an increased nitrate input from the roots. However, measurements of other available nitrogen forms would be necessary to convincingly rule out if increased nitrogen input has a role in willow phytoremediation. Indeed, nitrogen transfer from willow to the rhizosphere microorganisms might take the form of amino acids, and gene expression related to their uptake and utilization were highly elevated in the rhizosphere as compared to the bulk soil.
Increased expression of hydrocarbon degradation genes
In soils contaminated with organic pollutants, the goal of phytoremediation is to stimulate rhizobacteria to degrade the organic contaminants. Considering this, our 6-month pot experiment was successful in showing a generally higher expression of many hydrocarbon-degrading genes in the rhizosphere of willow. As expected, the expression of hydrocarbon degradation genes was mainly governed by soil contamination. However, within this overarching effect of contaminant presence, willows did have significant effects on the expression of functional genes related to hydrocarbon degradation. Willows induced the expression of several genes that were expressed at very low levels in the contaminated bulk soil. One of the most interesting features of phytoremediation is that many plant-derived chemicals (root degradation compounds, secondary metabolites, etc.) stimulate microorganisms to degrade contaminants (Donnelly et al., 1994; Fletcher and Hegde, 1995; Haby and Crowley, 1996; Miya and Firestone, 2001; Isidorov and Jdanova, 2002). Many of these plant-derived compounds are PAH analogs (Singer et al., 2003), and the rhizosphere of plants growing in uncontaminated soils is already enriched in PAH-degrading microorganisms (Daane et al., 2001). Many rhizosphere organisms also express aromatic degradation genes even in the absence of aromatic contaminants in the soil. In the absence of contaminant, it has been shown that Pseudomonas aromatic-degrading genes are upregulated in the corn rhizosphere, and that mutation in these genes results in a reduced fitness to rhizosphere conditions (Matilla et al., 2007). In the rhizosphere of various plants growing in non-contaminated soils, the shikimate and protocatechuate transport systems of Rhizobium leguminosarum were induced, and, when these genes were mutated, it led to reduced efficiency in rhizosphere colonization (Ramachandran et al., 2011). When exposing Pseudomonas aeruginosa to root exudates, genes related to aromatic compound catabolism and protocatechuate 3,4-dioxygenase were significantly upregulated (Mark et al., 2005).
More specifically, several willow species are recognized for their production of salicylic acid and related compounds. Salicylate is derived from the amino acid phenylalanine and is degraded through the catechol or gentisate pathways (Ishiyama et al., 2004). Salicylate-degrading bacteria were previously found to be enriched beneath willows in comparison with other vegetation (Schmidt et al., 2000). Salicylate has been shown to induce transcription of genes involved in the degradation of PAH (Schell, 1985) and polychlorinated biphenyl (Master and Mohn, 2001), to enhance the microbial degradation of naphthalene (Yen and Gunsalus, 1982; Van Der Meer et al., 1992) and other PAHs, such as fluoranthene, pyrene, benz[a]anthracene, chrysene and benz[a]pyrene (Chen and Aitken, 1999), and to function as a growth substrate for polychlorinated biphenyl-degrading bacteria (Singer et al., 2003). Here, within a complex microbial community, many PAH-degrading genes were significantly more expressed in the rhizosphere of willows planted in contaminated soils. More tests would be necessary to discriminate the role of salicylate among other possible mechanisms explaining this increased gene expression. If salicylate is confirmed as having a major role in enhancing the survival of PAH-degrading microorganisms and increasing PAH degradation activities, improved phytoremediation could be achieved by breeding willow cultivars that have increased production of salicylate. Some researchers have investigated the efficacy of amending PAH-contaminated soil with salicylate in an effort to induce PAH degradation by indigenous soil microorganisms (Ogunseitan et al., 1991; Colbert et al., 1993).
The major weakness of phytoremediation is that the effect of plants is largely limited to the rhizosphere, and, as a consequence, phytoremediation treatments typically proceed at slower rates than other bioremediation techniques. Even with fast-growing deep-rooting trees such as willows, phytoremediation treatment during this 6-month experiment did not significantly increase global hydrocarbon degradation in the pots, as the final hydrocarbon concentrations were not significantly different between the planted and unplanted pots. The probable cause is that sufficient rhizosphere soil was not available for chemical analyses, thus soil outside the direct influence of roots (bulk soil) was included in samples for the determination of hydrocarbon concentrations. However, the fact that the contamination levels did not differ significantly at the end of our study was an added benefit, as it allowed us to discriminate between the effect of willow and contamination, without having the confounding effect of different contamination levels in planted and unplanted pots.
Conclusions
This study provided the first in-depth metatranscriptomic view of microbial gene expression in the rhizosphere. Our results indicate a strong rhizosphere effect, with increased expression of many genes related to carbon and amino-acid utilization, nitrogen cycling and hydrocarbon degradation. This rhizosphere effect was in many cases more pronounced in contaminated soils. The large increase in the expression of hydrocarbon degradation genes in the rhizosphere of willow is promising, even though after only 6 months, we did not observe a significant effect of willows on hydrocarbon concentration outside the rhizosphere compartment. Several potential mechanisms explaining the stimulation of rhizosphere microbes were suggested: increased nitrogen inputs, increased plant exudation due to stressful conditions and exudation of salicylate and other PAH analogs. Future studies will be focused on precisely determining the mechanism behind the observed rhizosphere stimulation of hydrocarbon degraders and devising potential ways to improve phytoremediation.
References
Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM . (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57: 233–266.
Barac T, Taghavi S, Borremans B, Provoost A, Oeyen L, Colpaert JV et al (2004). Engineered endophytic bacteria improve phytoremediation of water-soluble, volatile, organic pollutants. Nat Biotechnol 22: 583–588.
Barnard R, Leadley PW, Hungate BA . (2005). Global change, nitrification, and denitrification: a review. Glob Biogeochem Cy 19: GB1007.
Bell TH, El-Din Hassan S, Lauron-Moreau A, Al-Otaibi F, Hijri M, Yergeau E et al (2014). Linkage between bacterial and fungal rhizosphere communities in hydrocarbon-contaminated soils is related to plant phylogeny. ISME J 8: 331–343.
Bell TH, Yergeau E, Maynard C, Juck D, Whyte LG, Greer CW . (2013). Predictable bacterial composition and hydrocarbon degradation in Arctic soils following diesel and nutrient disturbance. ISME J 7: 1200–1210.
Berg G, Roskot N, Steidle A, Eberl L, Zock A, Smalla K . (2002). Plant-dependent genotypic and phenotypic diversity of antagonistic rhizobacteria isolated from different Verticillium host plants. Appl Environ Microbiol 68: 3328–3338.
Berg G, Zachow C, Lottmann J, Gotz M, Costa R, Smalla K . (2005). Impact of plant species and site on rhizosphere-associated fungi antagonistic to Verticillium dahliae Kleb. Appl Environ Microbiol 71: 4203–4213.
Chen S-H, Aitken MD . (1999). Salicylate stimulates the degradation of high-molecular weight polycyclic aromatic hydrocarbons by Pseudomonas saccharophila P15. Environ Sci Technol 33: 435–439.
Cohen SP, Levy SB, Foulds J, Rosner JL . (1993). Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J Bacteriol 175: 7856–7862.
Colbert SF, Isakeit T, Ferri M, Weinhold AR, Hendson M, Schroth MN . (1993). Use of an exotic carbon source to selectively increase metabolic activity and growth of Pseudomonas putida in soil. Appl Environ Microbiol 59: 2056–2063.
Costerton JW, Stewart PS, Greenberg EP . (1999). Bacterial biofilms: A common cause of persistent infections. Science 284: 1318–1322.
Daane L, Harjono I, Zylstra G, Häggblom M . (2001). Isolation and characterization of polycyclic aromatic hydrocarbon-degrading bacteria associated with the rhizosphere of salt marsh plants. Appl Environ Microbiol 67: 2683–2691.
De Carcer DA, Martin M, Karlson U, Rivilla R . (2007a). Changes in bacterial populations and in biphenyl dioxygenase gene diversity in a polychlorinated biphenyl-polluted soil after introduction of willow trees for rhizoremediation. Appl Environ Microbiol 73: 6224–6232.
De Carcer DA, Martin M, Mackova M, Macek T, Karlson U, Rivilla R . (2007b). The introduction of genetically modified microorganisms designed for rhizoremediation induces changes on native bacteria in the rhizosphere but not in the surrounding soil. ISME J 1: 215–223.
De Ridder-Duine AS, Kowalchuk GA, Gunnewiek P, Smant W, van Veen JA, de Boer W . (2005). Rhizosphere bacterial community composition in natural stands of Carex arenaria (sand sedge) is determined by bulk soil community composition. Soil Biol Biochem 37: 349–357.
Dennis PG, Miller AJ, Hirsch PR . (2010). Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol Ecol 72: 313–327.
Diehn M, Relman DA . (2001). Comparing functional genomic datasets: lessons from DNA microarray analyses of host-pathogen interactions. Curr Opin Microbiol 4: 95–101.
Donnelly PK, Hegde RS, Fletcher JS . (1994). Growth of PCB-degrading bacteria on compounds from photosynthetic plants. Chemosphere 28: 981–988.
Esperschutz J, Pritsch K, Gattinger A, Welzl G, Haesler F, Buegger F et al (2009). Influence of chronic ozone stress on carbon translocation pattern into rhizosphere microbial communities of beech trees (Fagus sylvatica L.) during a growing season. Plant Soil 323: 85–95.
Fan B, Carvalhais L, Becker A, Fedoseyenko D, von Wiren N, Borriss R . (2012). Transcriptomic profiling of Bacillus amyloliquefaciens FZB42 in response to maize root exudates. BMC Microbiol 12: 116.
Feng Y-S, Lee C-M . (2009). The potential of the acetonitrile biodegradation by Mesorhizobium sp. F28. J Haz Mat 164: 646–650.
Fletcher JS, Hegde RS . (1995). Release of phenols by perennial plant roots and their potential importance in bioremediation. Chemosphere 31: 3009–3016.
Glass DJ . (1999) U.S. and International Markets for Phytoremediation, 1999–2000. D. Glass Associates: Needham: MA, USA., pp 270.
Gomes NCM, Fagbola O, Costa R, Rumjanek NG, Buchner A, Mendona-Hagler L et al (2003). Dynamics of fungal communities in bulk and maize rhizosphere soil in the tropics. Appl Environ Microbiol 69: 3758–3766.
Gonzalez-Pasayo R, Martinez-Romero E . (2000). Multiresistance Genes of Rhizobium etli CFN42. Mol Plant-Microbe Interact 13: 572–577.
Griffiths RI, Bailey MJ, McNamara NP, Whiteley AS . (2006). The functions and components of the Sourhope soil microbiota. Appl Soil Ecol 33: 114–126.
Haby PA, Crowley DE . (1996). Biodegradation of 3-chlorobenzoate as affected by rhizodeposition and selected carbon substrates. J Environ Qual 25: 304–310.
Haichar F, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent J et al (2008). Plant host habitat and root exudates shape soil bacterial community structure. ISME J 2: 1221–1230.
Hamady M, Lozupone C, Knight R . (2010). Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4: 17–27.
Han J-I, Choi H-K, Lee S-W, Orwin PM, Kim J, LaRoe SL et al (2011). Complete genome sequence of the metabolically versatile plant growth-promoting endophyte Variovorax paradoxus S110. J Bacteriol 193: 1183–1190.
Henry A, Doucette W, Norton J, Bugbee B . (2007). Changes in crested wheatgrass root exudation caused by flood, drought, and nutrient stress. J Environ Qual 36: 904–912.
Henry S, Texier S, Hallet S, Bru D, Dambreville C, Cheneby D et al (2008). Disentangling the rhizosphere effect on nitrate reducers and denitrifiers: insight into the role of root exudates. Environ Microbiol 10: 3082–3092.
Hogan D, Kolter R . (2002). Why are bacteria refractory to antimicrobials? Curr Opin Microbiol 5: 472–477.
Hrynkiewicz K, Ciesielska A, Haug I, Baum C . (2009). Ectomycorrhiza formation and willow growth promotion as affected by associated bacteria: role of microbial metabolites and use of C sources. Biol Fertil Soils 46: 139–150.
Huang A, Teplitski M, Rathinasabapathi B, Ma L . (2010). Characterization of arsenic-resistant bacteria from the rhizosphere of arsenic hyperaccumulator Pteris vittata. Can J Microbiol 56: 236–246.
Ishiyama D, Vujaklija D, Davies J . (2004). Novel pathway of salicylate degradation by Streptomyces sp. Strain WA46. Appl Environ Microbiol 70: 1297–1306.
Isidorov V, Jdanova M . (2002). Volatile organic compounds from leaves litter. Chemosphere 48: 975–979.
Jones DL . (1998). Organic acids in the rhizosphere - a critical review. Plant Soil 205: 25–44.
Jones DL, Hodge A, Kuzyakov Y . (2004). Plant and mycorrhizal regulation of rhizodeposition. New Phytol 163: 459–480.
Kielak A, Pijl AS, van Veen JA, Kowalchuk GA . (2008). Differences in vegetation composition and plant species identity lead to only minor changes in soil-borne microbial communities in a former arable field. FEMS Microbiol Ecol 63: 372–382.
Kowalchuk GA, Buma DS, de Boer W, Klinkhamer PGL, van Veen JA . (2002). Effects of above-ground plant species composition and diversity on the diversity of soil-borne microorganisms. Antonie Van Leeuwenhoek 81: 509–520.
Kowalchuk GA, Yergeau E, Leveau JHJ, Sessitch A, Bailey M . (2010). Plant-associated microbial communities. In: Liu W-T, Jansson JK (eds) Environmental Molecular Microbiology. Caister Academic Press: Norwich, UK, pp 133–147.
Kuffner M, Puschenreiter M, Wieshammer G, Gorfer M, Sessitsch A . (2008). Rhizosphere bacteria affect growth and metal uptake of heavy metal accumulating willows. Plant Soil 304: 35–44.
Kuiper I, Lagendijk EL, Bloemberg GV, Lugtenberg BJJ . (2004). Rhizoremediation: A Beneficial Plant-Microbe Interaction. Mol Plant-Microbe Interact 17: 6–15.
Leigh MB, Prouzova P, Mackova M, Macek T, Nagle DP, Fletcher JS . (2006). Polychlorinated biphenyl (PCB)-degrading bacteria associated with trees in a PCB-contaminated site. Appl Environ Microbiol 72: 2331–2342.
Lugtenberg BJJ, Dekkers L, Bloemberg GV . (2001). Molecular determinants of rhizosphere colonization by Pseudomonas. Annu Rev Phytopathol 39: 461–490.
Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S et al (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488: 86–90.
Marilley L, Aragno M . (1999). Phylogenetic diversity of bacterial communities differing in degree of proximity of Lolium perenne and Trifolium repens roots. Appl Soil Ecol 13: 127–136.
Mark GL, Dow JM, Kiely PD, Higgins H, Haynes J, Baysse C et al (2005). Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe-plant interactions. Proc Natl Acad Sci USA 102: 17454–17459.
Master ER, Mohn WW . (2001). Induction of bphA, encoding biphenyl dioxygenase, in two polychlorinated biphenyl-degrading bacteria, psychrotolerant Pseudomonas strain Cam-1 and MesophilicBurkholderia strain LB400. Appl Environ Microbiol 67: 2669–2676.
Mathesius U, Mulders S, Gao M, Teplitski M, Caetano-Anollés G, Rolfe BG et al (2003). Extensive and specific responses of a eukaryote to bacterial quorum-sensing signals. Proc Natl Acad Sci USA 100: 1444–1449.
Matilla M, Espinosa-Urgel M, Rodriguez-Herva J, Ramos J, Ramos-Gonzalez M . (2007). Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere. Genome Biol 8: R179.
Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M et al (2008). The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9: 386.
Meyer M, Kircher M . (2010). Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc doi:10.1101/pdb.prot5448.
Miya RK, Firestone MK . (2001). Enhanced phenanthrene biodegradation in soil by slender oat root exudates and root debris. J Environ Qual 30: 1911–1918.
Naik D, Smith E, Cumming JR . (2009). Rhizosphere carbon deposition, oxidative stress and nutritional changes in two poplar species exposed to aluminum. Tree Physiol 29: 423–436.
Nakatsu CH, Hristova K, Hanada S, Meng X-Y, Hanson JR, Scow KM et al (2006). Methylibium petroleiphilum gen. nov., sp. nov., a novel methyl tert-butyl ether-degrading methylotroph of the Betaproteobacteria. Int J Syst Evol Microbiol 56: 983–989.
Newsholme C . (2003) Willows: The Genus Salix. Timber Press, Incorporated: Portland, OR, USA, pp 256.
Ogunseitan OA, Delgado IL, Tsai YL, Olson BH . (1991). Effect of 2-hydroxybenzoate on the maintenance of naphthalene-degrading pseudomonads in seeded and unseeded soil. Appl Environ Microbiol 57: 2873–2879.
Pilon-Smits E . (2005). Phytoremediation. Ann Rev Plant Biol 56: 15–39.
Pulford I, Watson C . (2003). Phytoremediation of heavy metal-contaminated land by trees—a review. Environ Int 29: 529–540.
Qin R, Hirano Y, Brunner I . (2007). Exudation of organic acid anions from poplar roots after exposure to Al, Cu and Zn. Tree Physiol 27: 313–320.
Ramachandran V, East A, Karunakaran R, Downie JA, Poole P . (2011). Adaptation of Rhizobium leguminosarum to pea, alfalfa and sugar beet rhizospheres investigated by comparative transcriptomics. Genome Biol 12: R106.
Ramey BE, Koutsoudis M, SBv Bodman, Fuqua C . (2004). Biofilm formation in plant-microbe associations. Curr Opin Microbiol 7: 602–609.
Rosario-Passapera R, Keddis R, Wong R, Lutz RA, Starovoytov V, Vetriani C . (2012). Parvibaculum hydrocarboniclasticum sp. nov., a mesophilic, alkane-oxidizing alphaproteobacterium isolated from a deep-sea hydrothermal vent on the East Pacific Rise. Int J Syst Evol Microbiol 62: 2921–2926.
Salt DE, Smith RD, Raskin I . (1998). Phytoremediation. Annu Rev Plant Physiol Plant Mol Biol 49: 643–668.
Schell MA . (1985). Transcriptional control of the nah and sal hydrocarbon-degradation operons by the nahR gene product. Gene 36: 301–309.
Schenk PM, Carvalhais LC, Kazan K . (2012). Unraveling plant-microbe interactions: can multi-species transcriptomics help? Trends Biotechnol 30: 177–184.
Schleheck D, Tindall BJ, Rossello-Mora R, Cook AM . (2004). Parvibaculum lavamentivorans gen. nov., sp. nov., a novel heterotroph that initiates catabolism of linear alkylbenzenesulfonate. Int J Syst Evol Microbiol 54: 1489–1497.
Schmidt SK, Lipson DA, Raab TK . (2000). Effects of willows (Salix brachycarpa) on populations of salicylate-mineralizing microorganisms in alpine soils. J Chem Ecol 26: 2049–2057.
Schnoor JL, Licht LA, McCutcheon SC, Lee Wolfe N, Carreira LH . (1995). Phytoremediation of organic and nutrient contaminants. Environ Sci Technol 29: 318A–323A.
Singer AC, Crowley DE, Thompson IP . (2003). Secondary plant metabolites in phytoremediation and biotransformation. Trends Biotechnol 21: 123–130.
Sipilä TP, Keskinen AK, Akerman ML, Fortelius C, Haahtela K, Yrjälä K . (2008). High aromatic ring-cleavage diversity in birch rhizosphere: PAH treatment-specific changes of IE3 group extradiol dioxygenases and 16S rRNA bacterial communities in soil. ISME J 2: 968–981.
Smalla K, Wieland G, Buchner A, Zock A, Parzy J, Kaiser S et al (2001). Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: Plant-dependent enrichment and seasonal shifts revealed. Appl Environ Microbiol 67: 4742–4751.
Stewart FJ, Ottesen EA, DeLong EF . (2010). Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME J 4: 896–907.
Streeter J, Wong PP . (1988). Inhibition of legume nodule formation and N2 fixation by nitrate. Crit Rev Plant Sci 7: 1–23.
Taghavi S, Barac T, Greenberg B, Borremans B, Vangronsveld J, van der Lelie D . (2005). Horizontal gene transfer to endogenous endophytic bacteria from poplar improves phytoremediation of toluene. Appl Environ Microbiol 71: 8500–8505.
Turner T, Poole P . (2013). Transcriptomics and metatranscriptomic analysis of the response of rhizosphere bacteria to environmental change. In de Bruijn FJ (ed). Molecular Microbial Ecology of the Rhizosphere. John Wiley & Sons, Inc.: Hoboken, NJ, USA, pp 1123–1128.
Turner TR, Ramakrishnan K, Walshaw J, Heavens D, Alston M, Swarbreck D et al (2013). Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. The ISME J 7: 2248–2258.
Van Der Meer JR, De Vos WM, Harayama S, Zehnder A . (1992). Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol Rev 56: 677–694.
Vidal C, Chantreuil C, Berge O, Maure L, Escarre J, Bena G et al (2009). Mesorhizobium metallidurans sp. nov., a metal-resistant symbiont of Anthyllis vulneraria growing on metallicolous soil in Languedoc, France. Int J Syst Evol Microbiol 59: 850–855.
Weyens N, Schellingen K, Beckers B, Janssen J, Ceulemans R, van der Lelie D et al (2013). Potential of willow and its genetically engineered associated bacteria to remediate mixed Cd and toluene contamination. J Soils Sediments 13: 176–188.
Yen K, Gunsalus I . (1982). Plasmid gene organization: naphthalene/salicylate oxidation. Proc Natl Acad Sci USA 79: 874–878.
Yergeau E, Lawrence JR, Sanschagrin S, Waiser MJ, Korber DR, Greer CW . (2012). Next-generation sequencing of microbial communities in the Athabasca River and its tributaries in relation to oil sands mining activities. Appl Environ Microbiol 78: 7626–7637.
Zimmer D, Baum C, Leinweber P, Hrynkiewicz K, Meissner R . (2009). Associated bacteria increase the phytoextraction of cadmium and zinc from a metal-contaminated soil by mycorrhizal willows. Int J Phytorem 11: 200–213.
Acknowledgements
Werther Guidi, Michel Labrecque, Benoît Cloutier-Hurteau, Marie-Claude Turmel and François Courchesne provided soil analysis data, Yves Terrat and Sébastien Halary helped with soil sampling and Danielle Ouellette performed the RT-quantitative PCR analyses. We are grateful to ConocoPhillips for providing us with access to the Varennes field site. This project was supported by the Genome Canada and Genome Québec funded GenoRem Project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The GenoRem project contains several industrial partners, including ConocoPhillips, the company that provided us with access to their site for this study. Our manuscript has in no way been modified by ConocoPhillips, nor has any industrial partner commented on, or had any influence in, the analysis of our results. The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Rights and permissions
About this article
Cite this article
Yergeau, E., Sanschagrin, S., Maynard, C. et al. Microbial expression profiles in the rhizosphere of willows depend on soil contamination. ISME J 8, 344–358 (2014). https://doi.org/10.1038/ismej.2013.163
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ismej.2013.163
Keywords
This article is cited by
-
Effects of willow and Sedum alfredii Hance planting patterns on phytoremediation efficiency under AC electric field
Environmental Science and Pollution Research (2023)
-
The Effects of Agronomic Herbaceous Plants on the Soil Structure of Gold Mine Tailings and the Establishment of Boreal Forest Tree Seedlings
Water, Air, & Soil Pollution (2022)
-
Trees, fungi and bacteria: tripartite metatranscriptomics of a root microbiome responding to soil contamination
Microbiome (2018)