Abstract
Molecular variance was estimated in seven populations of the endemic species Scalesia affinis within and among islands of the Galápagos. The analysis, based on 157 polymorphic AFLP markers, revealed a high differentiation among populations, of which most was partitioned among islands. In addition, the information content of AFLP markers was tested with sets of discriminant analyses based on different numbers of AFLP markers. This indicated that the markers were highly informative in discriminating the populations. Although one of four populations from the island Isabela was sampled from a volcano 100 km away from the remaining populations, this population resembled the others on Isabela. The partitioning of molecular variance (AFLP) resulted in two unities, one consisting of populations from Isabela and one of populations from Santa Cruz and Floreana. The differentiation in two chloroplast microsatellites was higher than for AFLP markers and equally partitioned among populations within islands as among islands. Thus, gene flow via fruits within islands is as limited as among islands. The lower differentiation within islands in the nuclear AFLP markers may thus indicate that gene flow within islands is mostly accounted for by pollen transfer. S. affinis is the only species in the genus that is not listed in 2000 IUCN Red List of Threatened Species. However, due to prominent grazing and land exploitation, some populations have recently been reduced markedly, which was reflected in lower diversity. As inbreeding depression is present in the species, the rapid bottlenecks are threats to the populations.
Similar content being viewed by others
Introduction
The biota of oceanic islands is highly valuable for studies of evolutionary processes and has earned attention from many evolutionary biologists (eg Carlquist, 1974; Wagner and Funk, 1995; Grant, 1998; Stuessy and Ono, 1998). The biota often differ from those on the adjacent mainlands and may thus act with different selective pressures. Furthermore, the islands are spatially isolated from the mainland and from other islands, which inhibits gene flow between the newly established populations and the population of origin. Thus, mutations that occur on islands may spread under different selection pressures and become fixed rapidly because of isolation, potentially resulting in speciation (Barton, 1998). Small population sizes are often associated with island colonization. When the colonizers arrive at a remote island, the new populations will be founded by only one or a few individuals, resulting in a higher likelihood of allele fixation (Mayr, 1954). If the mainland population is distant from the island, the probability of receiving more conspecific individuals is small. The bottleneck caused by island colonization may lead to fixation of deleterious, alleles, thus resulting in inbreeding depression (Frankham, 1995; Lande, 1999). Island populations may hence be particularly vulnerable to go extinct because of inbreeding depression (Frankham, 1998).
The patchy spatial distribution of island populations will influence the amount and distribution of genetic diversity among the populations. If gene flow is restricted among subpopulations, genetic subdivision will increase through an increase in the number of matings between closely related individuals (Wright, 1938, 1951). Among patchy populations, as island populations are assumed to be, gene flow is expected to decrease with increasing distance between populations whereas genetic differentiation should rise because of genetic drift and or differing selection pressures (Wright, 1943; Kimura and Weiss, 1964; Slatkin, 1993).
Regarding the flora of the Galápagos, the largest endemic genus, Scalesia Arn., is the best example of a plant group that has undergone diversification. According to the most recent monograph of the genus, it comprises 14 species, four subspecies and two varieties (Eliasson, 1974). In 1986 an additional species was described (S. gordilloi Hamann and Wium-Andersen, 1986). The genus is scattered throughout the archipelago and representatives are found on almost all larger islands. The differentiation of the genus is particularly evident in leaf morphology. The variation in leaf morphology goes beyond the variation characteristic of some of the largest genera among the relatives of Scalesia that are found on the mainland (Howell, 1941). Most of the species have very limited distribution ranges; some are even limited to a few populations on a single island. One of the widely distributed species is S. affinis Hook. f., which occurs on the islands Isabela, Fernandina, Santa Cruz, and Floreana. The largest island Isabela comprises a chain of six volcanoes. Barren lava fields separate the four northern ones from each other and from southern Isabela (Willerslev et al, 2002). The biota on these volcanoes is distinct and it has been suggested that the volcanoes have always been separated from each other and from the southern part by either lava or sea Geist (1996). Hence, in biogeographical studies the four northern volcanoes have been treated as separate islands (Willerslev et al, 2002). S. affinis is found on all volcanoes of Isabela.
Due to both natural circumstances and human interference, the species of Scalesia are exposed to several threats. The archipelago is volcanic in origin and eruption still occurs and may further cause natural fires. The most severe threats are, however, land use for either agricultural or housing purposes (Adsersen, 1989). Other threatening factors are herbivory caused by introduced mammals such as goats, pigs and donkeys and the introduction of foreign plants (Adsersen, 1989; Mauchamp et al, 1998). S. affinis is the only species of Scalesia that is not listed in the IUCN Red List of Threatened Species (Hilton-Taylor, 2000). However, in recent years the populations on Santa Cruz and Floreana have been highly reduced as a result of both land exploitation and grazing (Adsersen, pers. comm.), and it was suggested by Itow (1995) that the populations of S. affinis on these islands should be considered vulnerable.
Here I analyse the differentiation of populations of S. affinis within and among islands of the Galápagos by using both biparentally (AFLP) and maternally inherited (two chloroplast microsatellites) markers and address the following questions. How does the partitioning of molecular variance fit to a landscape defined by volcanoes and islands? Is gene flow among populations predominantly accounted for by fruits (in S. affinis the one-seeded dispersal units, which the mature capitulum is composed of) or by pollen? The taxonomic status of the populations in the archipelago has been debated (eg Howell, 1941; Harling, 1962; Eliasson, 1974). What does molecular data tell about the taxonomy of S. affinis s. l.? In addition, I test the information content of AFLP markers by performing analyses with an increasing number of markers.
Materials and methods
Plant species
S. affinis is an evergreen perennial shrub 3 m tall. It occurs on lava beds from sea level to approximately 1000 m. The species is tetraploid (2n=68) as all other examined species of Scalesia (Eliasson, 1974).
Distribution
S. affinis occurs on the islands Fernandina, Isabela, Floreana and Santa Cruz. On Fernandina it is found mainly near sea level, but occurs also on the north-eastern slope at elevations between 900 and 1000 m (Eliasson, 1974). S. affinis is relatively common on southern Isabela, where it is found at scattered localities, mainly at low altitudes (Eliasson, 1974). On central and northern Isabela scattered populations are found in the vicinity of the four volcanoes up to ca. 500 m above sea level (Eliasson, 1974). S. affinis does not occur on the lava fields between the volcanoes. In 1974, S. affinis was relatively common at sea level on southern Santa Cruz and common on the western side of Floreana (Eliasson, 1974). In the years 1998–2001 where sampling for this study took place, only five individuals could be located on southern Santa Cruz despite intensive search. On western Floreana, S. affinis was still relatively abundant north of Black Beach (∼100 individuals), whereas only few individuals were found south of Black Beach.
Sampling
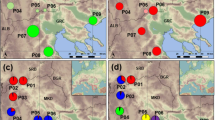
Leaf material was collected from seven localities as shown in Figure 1. I refer to the sampling localities as populations although the localities in southern Isabela may not have been completely spatially separated. Young leaves were stored in silica gel for DNA analysis. See Table 1 for sample sizes from each locality. On Santa Cruz all available individuals were sampled, at the other localities the individuals were randomly chosen. On Floreana samples were taken from two localities north and south of the village, respectively. At one of these localities, the angle and distance to a fixed point were registered for each individual for a later analysis of the spatial genetic structure within the population (Pop. 7).
Molecular data
Silica dried leaf material (15 mg) was used to extract genomic DNA using DNAeasy Plant Mini Kit (Qiagen) following supplied instructions with the following minor modifications. Between steps 4 and 5, the suggested option for very viscous lysates was applied. The preceding step was carried out with the supernatants after centrifuging the samples for 5 min at 13 000 rpm. At steps 8 and 9, extra centrifugations were added when necessary.
Amplified fragment length polymorphism (AFLP)
The AFLP technique was performed on 500 ng genomic DNA with the PE Applied Biosystems AFLPTM plant mapping kit for average-sized genomes supplemented by a Tamra-labelled selective primer from TAGCopenhagen A/S. The AFLP procedure followed the manufacturer's protocol. The method is based on Vos et al (1995), but the primers are labelled with nonradioactive fluorescent dye. In all, 28 selective primer combinations were tested, of which four were chosen that gave good amplifications and that showed polymorphisms. The combinations were EcoRI(Fam)-ACT/MseI-CAG, EcoRI(Joe)-AGG/MseI-CTC, EcoRI(Tamra)-ACC/MseI-CAC and EcoRI(Fam)-ACT/MseI-CTC. Amplification was conducted on a PTC 200 Peltier Thermal Cycler (MJ Research). AFLP fragments were separated on an ABI Prism 377 sequencer using 5% denaturing Long Ranger acrylamide gels. An internal size standard (GS-500 ROX, PE Applied Biosystems) was included in each sample to facilitate fragment sizing and GeneScan Analysis Software 2.1 was used to estimate fragment size (PE Applied Biosystems, 1996a). Hereafter the GeneScan files were scored with Genotyper 2.0 (PE Applied Biosystems, 1996b) and a binary scoring table was constructed and exported to Microsoft Excel. Finally, each AFLP profile was visually checked by eye. Markers that failed in displaying good amplification in any of the individuals were excluded for further analyses. Fragments that differed by more than 0.5 relative migration units (bp) between samples were identified as different.
As the SAS procedures exclude individuals with missing values, missing values (0.23%) were replaced by simulated values before analysing the data. A 1 substituted a missing value if a random number (uniformly distributed on the interval 0–1) was equal to or smaller than the frequency of that marker in the given population. Otherwise, a 0 substituted the value. One individual in the small population (Pop. 5) contained many missing values and was therefore excluded from most of the analyses. This matrix, obtained from polymorphic markers only, was also used for the following analyses carried out with Arlequin 2.000 (Schneider et al, 2000).
Two measures of diversity were estimated at the seven localities. These were the proportion of polymorphic markers (P) and the mean number of differences between all pairs of haplotypes in the given population, divided by the total number of markers.
A matrix of pairwise Euclidean distances was estimated as defined by Excoffier et al (1992). To estimate variance components at different hierarchical levels, an analysis of molecular variance (AMOVA) was applied based on the Euclidean distances. These hierarchies were: among individuals within populations, among populations within islands and among islands. As the statistics are not directly comparable to the traditional Wright's F-statistics (Wright, 1951) they are referred to as Φ-statistics (Excoffier et al, 1992). The significance of ΦST (among populations relative to the total population) was tested by 10 000 permutations of haplotypes among populations among islands. The significance of ΦSC (among populations within islands) was tested by 10 000 permutations of haplotypes among populations within islands, and the significance of ΦCT (among islands) was tested by 10 000 permutations of populations among islands.
The relationships among the populations were described with a tree based on Euclidean distances, where the among-population distances were corrected for the average within population distances. The tree was estimated with the program FITCH (Felsenstein, 1993).
To test whether it was possible to separate the populations on the basis of their banding patterns, the AFLP data were subjected to a nonparametric discriminant analysis. The analysis was based on the matrix based on Euclidean distances, and on the nearest neighbours method, with the classification evaluated by crossvalidation (SAS Institute, 1990).
It was further tested how effective the AFLP markers were in discriminating individuals of S. affinis, by conducting the discriminant analysis with an increasing number of markers. This was carried out on both island and population levels. First, a random sample of 10 markers was drawn and the discriminant analysis was then based on these markers. This was repeated 10 times and the mean±standard deviation (SD) was estimated. After a new randomization round, the analysis was again performed 10 times now with 20 markers and the mean±SD was calculated. This continued up to the use of 150 markers.
The spatial genetic structure within population Pop. 7 was analysed with the program SGS: Spatial Genetic Structure (Degen, 2000). All calculated statistics were mean genetic distances between all pairs of individuals belonging to a given spatial distance class. The number of distance classes as well as the size of the distance classes were chosen so that each class included at least 30 pairs of data points as recommended. This resulted in the choice of five distance classes each of the size 120 m. The genetic dissimilarity, Dij, between two individuals i and j was computed using the Tanimoto distance:

where wij is the number of common fragments of individuals i and j, and yi and yj equal the number of fragments, which only exist in individuals i or j, respectively. The distances Dij are constrained within the interval [0, 1] (Deichsel and Trampisch, 1985). To test any significant deviation from spatial random distribution of each calculated measure, a permutation test using Monte-Carlo simulations was applied (Manly, 1997). For each of the five distance classes, observed values were compared with the distribution obtained after 1000 permutations.
Chloroplast microsatellites
Amplification of two chloroplast microsatellite loci was completed with pairs of chloroplast microsatellite primers from TAGCopenhagen A/S previously designed for Scalesia divisa. The sequences of the microsatellite primers were (1) forward primer (trnT–trnL region) 5′-CAC GAT TAT TTT ATC ACG-3′, reverse primer 5′-TTC CAC TGA TTT TCC CCT-3′ and (2) forward primer (psbC–trnS region) 5′-CGG TTC CCT CAT ACA TAT AG-3′ and reverse primer 5′-CGC CTA GCC AAG CCA GAA A-3′. The forward primers were labelled with the fluorescent dyes Joe and Fam, respectively. PCR reactions of a total volume of 20 μl were prepared (2 μl 10 × Taq buffer incl. 0.2 mM MgCl2, 8 μl of a solution consisting of 0.5 mM of each dNTP, 2 μl of each primer (10 μM), 0.1 μl 5000 U/ml Taq DNA polymerase, 5 μl sterile MilliQ water and 1 μl template DNA). A PTC 200 Peltier Thermal Cycler (MJ Research) was used for amplification under the following profiles. Locus 1:4 min denaturing at 94°C, followed by 35 cycles of 30 s denaturing at 94°C, 1 min annealing at 47°C and 1 min extension at 72°C, with a final extension step at 72°C for 10 min. The amplification products varied in size from 91–103 bp. Locus 2: as Locus 1, but with the annealing temperature at 55°C. The length of the amplified products was from 92 to 98 bp.
The PCR products were visualized on a 2% agarose gel, and diluted according to the intensity of the bands. The diluted samples were combined with an internal size standard (GS-500 ROX labeled standard from PE Applied Biosystems) and run for 2 h on a 5% Long Ranger acrylamide gel on an ABI Prism 377 sequencer. Fragment sizing was facilitated with GeneScan Analysis Software 2.1 and hereafter analysed with Genotyper 2.0 (PE Applied Biosystems, 1996a, 1996b). For those populations that were not fixed in one or both chloroplast microsatellites, a test for linkage equilibrium was performed with Arlequin 2.000 (Schneider et al, 2000). The allele frequencies were subjected to an AMOVA with the same hierarchical structure as described above for the AFLP data.
Results
AFLP
There were a total of 286 interpretable markers. A marker was considered monomorphic if all individuals across all populations except for two (∼98%) showed the same genotype. According to this criterion, there were in total 129 monomorphic markers while the remaining 157 markers were considered polymorphic in at least one population.
In 30 of the 157 polymorphic markers individuals from the Isabela populations had banding patterns, which were not found in individuals from Floreana (eg a marker could be polymorphic among individuals on Isabela, while absent in individuals from Floreana). In 24 of these markers the population on Santa Cruz shared banding pattern with the Isabela populations, while the population resembled the Floreana populations in the remaining six markers.
The average proportion of polymorphic markers ranged from 0.414 in the small population on Santa Cruz to 0.803 in one of the populations from Floreana (Pop. 7, Table 1). The three populations on the southern part of Isabela Island had almost the same level, whereas the northern population had a slightly higher proportion. Regarding AFLP diversity, the small population on Santa Cruz had the lowest value (0.185±0.115), whereas the remaining populations were in the range from 0.200 (±0.101) to 0.248 (±0.126). The highest value was found in the population on northern Isabela (Pop. 4).
Table 2 shows that the pairwise differences among populations within the same island were clearly lower than among populations from different islands. This relationship is also reflected in the tree, where populations from the same island are grouped together (Figure 2). The small population from Santa Cruz was closer to the Floreana populations than to the populations from Isabela. Compared to the populations from Isabela, the Floreana populations appeared with relatively long branches indicating a relatively high differentiation.
The analysis of molecular variance revealed that the highest fraction of variation was found within populations (Table 3). The corresponding ΦST statistic was 0.435, indicating substantial differentiation among populations. This component was partitioned into two variance components: among populations within islands (ΦSC) and among populations among islands (ΦCT) and were 0.150 and 0.336, respectively (Table 3). Thus, most of the variation among populations was found among islands. All the three measures were significant with the following values P<0.001, P<0.001 and P=0.008, respectively.
No significance of a spatial genetic structure was found on short distances in the population analysed. However, as shown in Figure 3, the genetic dissimilarity increased significantly with the distance in metres at the distance class 240–360 m (P=0.005). At the following distance class, there was a similar tendency, though only close to significant (P=0.067). Hereafter no correlation was found.
The 157 polymorphic AFLP markers were highly informative. As shown in Table 4, four of the seven populations were so distinct that all individuals were classified into the population of origin without any errors. Misclassification only occurred among populations on Isabela. In addition, three individuals from Pop. 3, Pop. 4, and Pop. 2, respectively, had equal probabilities of assignment to two populations on Isabela. For each of these individuals, a coin was tossed to place the individuals randomly in one of possible populations. The perfect assignment of the populations on Floreana and Santa Cruz and the misclassifications on Isabela correspond with the relative lengths of the tree branches in Figure 2.
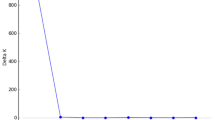
The error rate for classification is presented in Figures 4 and 5 for the discriminant analysis run with an increasing number of markers. The SD is clearly much higher when only few markers are included in the analyses. At the starting point (10 randomly chosen markers) the error rate is evidently lower when the analysis is based on islands (Figure 4) rather than populations (Figure 5). The islands are well discriminated by the use of 90 or more markers. When the analysis is population based, the error rate becomes stable at 10% when using 140 or more markers.
Chloroplast microsatellites
The allele frequencies of the two microsatellite loci in the seven populations are presented in Table 5. Linkage disequilibrium was found in two populations (Pop. 6 and Pop. 7) with P<0.001 and P=0.004, respectively. The general pattern was in agreement with the pattern obtained from AFLP markers. The three southern populations on Isabela Island resembled each other as they had the same high frequency of allele 2 in microsatellite locus 1 and were fixed for the same allele in locus 2. This allele is not found outside Isabela Island. The population on northern Isabela deviated from the southern ones by being fixed at both loci. Further, in microsatellite locus 1, the population was fixed for an allele that is common on Santa Cruz, while rare or absent in the remaining populations. The Santa Cruz population resembled the two Floreana populations most in locus 2, as allele 1 was the most common allele in all three populations. Further, Pop. 5 and Pop. 7 have an allele in common that is not found in any other population.
In Table 6 is given the results of the AMOVA when the microsatellites are treated as independent loci. Less than 20% of the total variation was found within populations, thus indicating a high among-population differentiation. This was equally partitioned within and among islands (ΦSC=0.549 and ΦCT=0.574, respectively). When the two loci are treated as one, the same fixation indices are ΦSC=0.657 and ΦCT=0.487, respectively, and thus comparable.
Discussion
All analyses indicate a high differentiation among the three islands. For instance, in the distance tree, the Isabela populations (Pop. 1, Pop. 2, Pop. 3, Pop. 4) are grouped together. The Floreana populations are likewise grouped together and are distant to the Isabela populations. The small population from Santa Cruz is located in between, however much closer to Floreana.
The differentiation found among populations in S. affinis was sufficient to assign most of the individuals correctly to their population of origin. Misassignment did not occur among islands; in fact misassignment was only found in a low rate among three populations on Isabela. When running the discriminant analysis with an increasing number of markers, the error rate became stable at 10% by the use of 140 or more markers. The error rate never reached 0, which is due to the relatively high similarity among the Isabela populations. The islands however were well discriminated by the use of 90 or more markers, which in S. affinis corresponds to three primer combinations.
The AMOVA based on AFLP markers likewise revealed significant differences among populations, representing approximately 44% of the total variance. When compared to 26 studies based on dominant markers (RAPD) listed by Bartish et al (1999), only six studies showed higher differentiation among populations than found in S. affinis. The high differentiation among populations in S. affinis corresponds well to what was found in another insular endemic plant, Campanula microdonta (Campanulaceae) from the Izu Islands (46%; Oiki et al, 2001). In C. microdonta, the differentiation was explained by the predominantly self-pollinating nature of the species combined with geographic isolation of the populations. S. affinis, however, is highly outcrossing (Nielsen et al, 2003) and the high differentiation must be due largely to a high among-island variation. When splitting the among-population differentiation in S. affinis into variation among populations within islands and among populations between islands, it is clear that most of the variation is found between islands (ΦSC=0.150 and ΦCT=0.336, respectively). The ocean between islands of the archipelago, hence, inhibits gene flow among populations located on different islands. The major pollinator of S. affinis is the endemic carpenter bee, Xylocopa darwini (Nielsen et al, 2003). Although it may be able to fly long distances, it seems unlikely that gene flow among islands is via pollen.
The high among-population differentiation found in the present study corresponds well with studies of insular taxa based on isozyme data. These studies have shown that island endemics tend to have a higher proportion of allozyme diversity among populations than endemic species in general (Francisco-Ortega et al, 2000; Crawford et al, 2001; Maki, 2001).
Compared to the AFLP differentiation, the chloroplast microsatellite variation showed an overall higher level of differentiation (Table 6). This was equally partitioned among populations within islands as among islands (ΦSC=0.549 and ΦCT=0.574, respectively). This may indicate that gene flow via fruits among populations within islands is as limited as among islands. The lower differentiation within islands in the nuclear AFLP markers thus shows that gene flow on islands is mostly accounted for by pollen transfer. The fruits of S. affinis are without structures that improve seed dispersal, such as hairiness or the presence of awns. S. affinis is however found on four islands in the archipelago, hence the fruits must occasionally disperse between islands. Documentation of seed dispersal of any Scalesia species is still lacking, but it has been suggested that seed-eating birds may have transferred the fruits from island to island (Carlquist, 1974; Elliason, 1974).
Parts of the high among-population differentiation found in the present study may result from human induced fragmentation of the natural populations. While the populations on Isabela constitute hundreds of members, the populations on both Santa Cruz and Floreana are very limited in size. The major threats to the populations are land exploitation and grazing by introduced mammals (Adsersen, 1989). While S. affinis was considered relatively common on Santa Cruz in 1974 (Eliasson, 1974), only five individuals could be located in 1998 when the sampling for this study took place. The population has thus been through a tremendous bottleneck over the last 30 years, which is also indicated by the lower number of polymorphic markers and low diversity relative to the other populations. Considering the populations on Floreana, these were surprisingly distinct although they were only separated spatially by a few kilometers (Figures 1 and 2). The population south of Black Beach (Floreana) was reduced from approximately 30 individuals in 1998 to about five individuals in 2000 (Henning Adsersen and Marianne Philipp, pers. comm.). There, donkeys are presumably the worst threat to S. affinis, biting of the capitula, thus preventing the maturation of new fruits. It is likely that the variation among populations on Floreana has been more continuous previously with and overall lower among-population differentiation than found in this study.
Like the AFLP characters, morphological characters have also shown differences that seem to be island related. Thus, the taxonomic status of the populations of S. affinis has been widely disputed over the years. Harling (1962) chose to divide S. affinis into three subspecies; ssp gummifera (Hook. f.) Harling, ssp affinis Hook. f. and ssp brachyloba Harling with the following distributions: ssp gummifera on Isabela and Fernandia, ssp affinis on Floreana and ssp brachyloba on Santa Cruz. In the latest revision, Eliasson (1974) believed in a closer alliance between specimens from Santa Cruz and Floreana than specimens from these two islands and specimens from Isabela and Fernandina. He was inclined to accept ssp gummifera and ssp affinis as classified by Harling. Eliasson (1974) further pointed out that the subspecies from Fernandina/Isabela show substantially more morphological variation than S. affinis s. str on Floreana. A plausible explanation for this pattern was the topography of Isabela with numerous volcanoes that harbour communities comparable to separate islands. In this study, material was included from the northern volcano Darwin as well as from the southern part of Isabela. The northern population was fixed at both microsatellite markers, which may have been caused by a bottleneck when the population was established. Regarding the AFLP markers, however, the population showed high similarity with the remaining Isabela populations even though it was separated from the populations in south by kilometres of lava fields with no vegetation. When comparing the partitioning of molecular variance (Figure 2) with the actual location of the populations in the archipelago (Figure 1), it is clear that the Isabela populations form one unity and the populations from Santa Cruz and Floreana another. Remembering the separation of northern and southern Isabela, it seems reasonable to suggest that the similarity of the four Isabela populations has a historic background. Thus, the present study supports high divergence between populations from Isabela and those from other islands, whereas the population from Santa Cruz shows high similarity with the two Floreana populations and may thus suggest that only two potential subspecies are supported; ssp gummifera and ssp affinis.
An obvious spatial genetic structure within populations is not as evident as the structure found on a larger scale. In the population analysed from Floreana no correlation was found between genetic dissimilarity among individuals and their spatial distance within the first 240 m (Figure 2). Hereafter the dissimilarity increased significantly with distance in the range 240–360 m (P=0.005) and close to significant (P=0.067) at 360–480 m. After this, no relationship appeared. The lack of genetic structure on shorter distances may reflect the outcrossing behaviour of the species. A previous study on the breeding system of S. affinis indicated high importance of the carpenter bee as pollinator (Nielsen et al, 2003). In all, 60% of the individuals produced 0–2 viable embryos after self-pollination, thus favouring outcrossing. Though geitonogamy was frequent in the population, the behaviour of the bees when visiting the observation site seemed to assure outcrossing. A recapture experiment showed that most of the bees (69%) visiting the observation patch were unmarked ‘new’ bees, thus suggesting a high degree of movement in the area (Nielsen et al, 2003). A high pollen transfer from plant to plant at shorter distances (up to approximately 250 m) seems thus to blur any substructure in the population.
It has been hypothesized that the progenitors of Scalesia reached the Galápagos Islands from South America by only one introduction (Schilling et al, 1994). As the oldest rocks of the Galápagos Islands have been estimated to be c. 4 Myr old (Cox, 1983), the entire evolution of Scalesia has taken place through a relatively short time. Where most of the described species of Scalesia have very small distribution areas, S. affinis is one of the two species of Scalesia that is found on several islands. The molecular variance of S. affinis revealed a patchy spatial distribution where the individuals were easily recognized at island level from their AFLP profile. In addition, the populations on Santa Cruz and Floreana have been through severe bottlenecks recently, with only a few individuals left in some of the populations. This may further increase genetic subdivision because the probability of biparental inbreeding increases (Wright, 1938, 1951). A further diversification may thus be facilitated during time.
Island populations may be particularly vulnerable to inbreeding depression because of fixation of deleterious alleles during colonization (Frankham, 1995, 1998; Lande, 1999). Nielsen et al (unpublished) found that S. affinis displayed a significant inbreeding depression when self-pollinated. Selfed offspring performed significantly worse than outcrossed offspring in several traits (eg germination success, survival, height growth, and leaf production) thus indicating prominent genetic load in the focal population. The species may thus be susceptible to rapid bottlenecks caused by herbivory and land use. S. affinis is the only species of Scalesia that is not listed in the 2000 IUCN Red List of Threatened Species (Hilton-Taylor, 2000). However, with continuous land exploitation and herbivory on Santa Cruz and Floreana, the populations of S. affinis on these two Islands will eventually become extinct.
References
Adsersen H (1989). The rare plants of the Galápagos Islands and their conservation. Biol Conserv 47: 49–77.
Bartish IV, Jeppsson N, Nybom H (1999). Population genetic structure in the dioecious pioneer plant species Hippophae rhamnoides investigated by random amplified polymorphic DNA (RAPD) markers. Mol Ecol 8: 791–802.
Barton NH (1998). Natural selection and random genetic drift as causes of evolution on islands. In: Grant PR (ed) Evolution on Islands, Oxford University Press: Oxford. pp 102–123.
Carlquist S (1974). Island Biology, Columbia Press: New York.
Cox A (1983). Ages of the Galápagos Islands. In: Bowman RI, Berson M, Leviton AE (eds) Patterns of Evolution in Galápagos Organisms, American Association for the Advancement of Science: San Francisco. pp 11–23.
Crawford DJ, Ruiz E, Stuessy TF, Tepe E, Aqeveque P, Gonzales F et al (2001). Allozyme diversity in endemic flowering plant species of the Juan Fernandez Archipelago, Chile: ecological and historical factors with implications for conservation. Am J Bot 88: 2195–2203.
Degen B (2000). SGS: Spatial Genetic Software. Computer program and user's manual http://kourou.cirad.fr/genetique/software.html.
Deichsel G, Trampisch HJ (1985). Clusteranalyse und Diskriminanzanalyse, Gustav Fischer Verlag: Stuttgart.
Eliasson U (1974). Studies in Galápagos Plants XIV. The Genus Scalesia Arn. Opera Bot 36: 1–117.
Excoffier L, Smouse PE, Zuattro JM (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479–491.
Felsenstein J (1993). PHYLIP (Phylogeny Inference Package) Version 3.5c. Distributed by the author, Department of Genetics, University of Washington.
Francisco-Ortega J, Santos-Guerra A, Kim S-C, Crawford DJ (2000). Plant genetic diversity in the Canary Islands: a conservation perspective. Am J Bot 87: 909–919.
Frankham R (1995). Inbreeding and extinction: a threshold effect. Conserv Biol 9: 792–799.
Frankham R (1998). Inbreeding and extinction: Island populations. Conserv Biol 12: 665–675.
Geist D (1996). On the emergence and submergence of the Galápagos Islands. Not Galápagos 56: 5–9.
Grant PR (ed) (1998). Evolution on Islands, Oxford University Press: Oxford.
Hamann O, Wium-Andersen S (1986). Scalesia gordilloi sp. Nov. (Asteraceae) from the Galápagos Islands, Ecuador. Nord J Bot 6: 35–38.
Harling G (1962). On some Compositae endemic to the Galápagos Islands. Acta Horti Bergiani 20: 63–120.
Hilton-Taylor C (compiler) (2000). 2000 IUCN Red List of Threatened Species, IUCN: Gland, Switzerland and Cambridge, UK.
Howell JT (1941). The genus Scalesia. Proc Calif Acad Sci Fourth Ser 22: 221–271.
Itow S (1995). Phytogeography and ecology of Scalesia (Compositae) endemic to the Galápagos Islands. Pac Sci 49: 17–30.
Kimura M, Weiss GH (1964). The stepping stone model of population structure and the decrease of genetic correlation with distance. Genetics 49: 561–576.
Lande R (1999). Extinction risks from anthropogenic, ecological, and genetic factors. In: Landweber LF, Dobson SP (eds) Genetics and Extinction of Species, Princeton University Press: Princeton. pp 1–22.
Maki M (2001). Genetic differentiation within and among island populations of the endangered plant Aster miyagii (Asteraceae), an endemic to the Ryukyu Islands. Am J Bot 88: 2189–2194.
Manly BFJ (1997). Randomization, Bootstrap and Monte Carlo Methods in Biology, Chapman & Hall: London.
Mauchamp A, Aldaz I, Edwin O, Valdebenito H (1998). Threatened species, a re-evaluation of the status of eight endemic plants of the Galápagos. Biodiv Conserv 7: 97–107.
Mayr E (1954). Change of genetic environment and evolution. In: Huxley J, Hardy AC, Ford EB (eds) Evolution as a Process, Allen and Unwin: London. pp 157–180.
Nielsen LR, Siegismund HR, Philipp M (2003). Partial self-incompatibility in the polyploid endemic species Scalesia affinis (Asteraceae) from the Galápagos: remnants of a self-incompatibility system? Bot J Linn Soc 142: 93–101.
Oiki S, Kawahara T, Inoue K, Ohara M, Maki M (2001). Random amplified polymorphic DNA (RAPD) variation among populations of the insular endemic plant Campanula microdonta (Campanulaceae). Ann Bot 87: 661–667.
Perkin-Elmer, Applied Biosystems (1996a). ABI Prism GeneScan Analysis Software (User's manual).
Perkin-Elmer, Applied Biosystems (1996b). ABI Prism Genotyper 2.0. (User's manual).
SAS (1990). SAS/STAT® User's Guide, Version 6, Vol 1, 4th edn. SAS Institute Inc.: Cary, NC.
Schilling EE, Panero JL, Eliasson UH (1994). Evidence from chloroplast DNA restriction site analysis on the relationships of Scalesia (Asteraceae: Heliantheae). Am J Bot 81: 248–254.
Schneider S, Roessli D, Excoffier L (2000). A Software for Population Genetics Data Analysis, Arlequin version 2.000. Genetics and Biometry Laboratory, University of Geneva, Switzerland.
Slatkin M (1993). Isolation by distance in equilibrium and non-equilibrium populations. Evolution 47: 264–279.
Stuessy TF, Ono M (eds) (1998). Evolution and Speciation of Island Plants, Cambridge University Press: Cambridge.
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M et al (1995). AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23: 4407–4414.
Willerslev E, Hansen AJ, Nielsen KK, Adsersen H (2002). Number of endemic and native plant species in the Galápagos Archipelago in relation to geographical parameters. Ecography 25: 109–119.
Wagner WL, Funk VA (eds) (1995). Hawaiian Biogeography. Evolution on a Hot Spot Archipelago, Smithsonian Institute Press: Washington.
Wright S (1938). Size of population and breeding structure in relation to evolution. Science 87: 430–431.
Wright S (1943). Isolation-by-distance. Genetics 28: 114–138.
Wright S (1951). The genetical structure of populations. Ann Eugen 15: 323–354.
Acknowledgements
The Charles Darwin Research Station is acknowledged for support during several field trips to the Galápagos Islands, and the Galápagos National Park Service is acknowledged for the permission given to the project. Henning Adsersen, Iván Aldaz, Marianne Philipp and Hans R Siegismund are thanked for collecting plant material from several localities. Hans R Siegismund, Henning Adsersen, Gitte Petersen and Marianne Philipp are further thanked for valuable comments on the manuscript. I am grateful to Michael Fay and Robyn Cowan (Royal Botanic Gardens, Kew) for their contribution to the technical part of this research. Ruth Bruus Jakobsen, Charlotte Hansen and Lisbeth Knudsen provided additional technical assistance. Financial support was received from The Danish Natural Science Research Council and from the Carlsberg Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nielsen, L. Molecular differentiation within and among island populations of the endemic plant Scalesia affinis (Asteraceae) from the Galápagos Islands. Heredity 93, 434–442 (2004). https://doi.org/10.1038/sj.hdy.6800520
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800520
Keywords
This article is cited by
-
Host-parasite ecology, behavior and genetics: a review of the introduced fly parasite Philornis downsi and its Darwin’s finch hosts
BMC Zoology (2016)
-
Exploring the legacy of goat grazing: signatures of habitat fragmentation on genetic patterns of endemic weevil populations in Northern Isabela Island, Galápagos (Ecuador)
Conservation Genetics (2016)
-
Genetic structures of Calophyllum inophyllum L., a tree employing sea-drift seed dispersal in the northern extreme of its distribution
Annals of Forest Science (2014)
-
Conservation genetics of an endemic from the Mediterranean Basin: high genetic differentiation but no genetic diversity loss from the last populations of the Sicilian Grape Hyacinth Leopoldia gussonei
Conservation Genetics (2013)
-
Population genetic structure and conservation of the Azorean tree Prunus azorica (Rosaceae)
Plant Systematics and Evolution (2013)