Abstract
The genetic architecture underlying nematode resistance and body weight in Blackface lambs was evaluated comparing genome-wide association (GWA) and regional heritability mapping (RHM) approaches. The traits analysed were faecal egg count (FEC) and immunoglobulin A activity against third-stage larvae from Teladorsagia circumcincta, as indicators of nematode resistance, and body weight in a population of 752 Scottish Blackface lambs, genotyped with the 50k single-nucleotide polymorphism (SNP) chip. FEC for both Nematodirus and Strongyles nematodes (excluding Nematodirus), as well as body weight were collected at approximately 16, 20 and 24 weeks of age. In addition, a weighted average animal effect was estimated for both FEC and body weight traits. After quality control, 44 388 SNPs were available for the GWA analysis and 42 841 for the RHM, which utilises only mapped SNPs. The same fixed effects were used in both analyses: sex, year, management group, litter size and age of dam, with day of birth as covariate. Some genomic regions of interest for both nematode resistance and body weight traits were identified, using both GWA and RHM approaches. For both methods, strong evidence for association was found on chromosome 14 for Nematodirus average animal effect, chromosome 6 for Strongyles FEC at 16 weeks and chromosome 6 for body weight at 16 weeks. Across the entire data set, RHM identified more regions reaching the suggestive level than GWA, suggesting that RHM is capable of capturing some of the variation not detected by GWA analyses.
Similar content being viewed by others
Introduction
Gastrointestinal nematode infections have a great impact on the sheep industry. Firstly, there is a direct annual cost of anthelmintic treatment and increased labour for sheep and pasture management, and thereafter production losses resulting from infections. Costs arising from these components have been quantified for the UK sheep to be approaching £100 million per year (Nieuwhof and Bishop, 2005). Control is largely based around anthelmintic usage; however, the development of anthelmintic resistance in nematode populations has fast become an international problem (Besier and Love, 2003; Bartley et al., 2004). This has led to the need for new control measures, such as selection for increased nematode resistance, with available field data (for example, Kemper et al., 2010), suggesting that such selection should be successful. However, the mechanisms underlying genetic differences in resistance are poorly categorised, with nematode resistance being a physiologically complex trait that develops over time. Therefore, indicator traits measured at any point in time might fail to represent all of the pathways involved in nematode resistance.
Traditionally, the genetic control of complex traits in livestock has been studied without identifying the genes or gene variants underlying observed variation, with selection on the basis of estimated breeding values calculated from phenotypic and pedigree information (Goddard and Hayes, 2009). Moreover, selection for nematode resistance has mainly been on the basis of use of indicator traits, such as faecal egg count (FEC) (for example, Morris et al., 1998; Bishop and Stear, 2001), with immunoglobulin A (IgA) activity (Coltman et al., 2001) also considered as an indicator of host response. However, collecting and quantifying such indicator traits is a costly and time-consuming process and usually requires the animal to undergo parasitic challenge. Therefore, it would be advantageous to select directly for parasite resistance without the requirement for challenge; current molecular tools allow animal geneticists to investigate the nature of genetic variation underlying complex traits, and in some circumstances this makes direct selection a possibility.
Several studies have reported quantitative trait loci (QTL) for nematode resistance in sheep (for example, Diez-Tascon et al., 2002; Crawford et al., 2006; Davies et al., 2006; Sallé et al., 2012). However, little overall consensus has emerged from these studies, an outcome compounded by the genetic complexity of the trait and the fact that these studies are very diverse, involving a variety of sheep breeds, nematode species and experimental approaches. Recently, however, genome-wide association (GWA) studies have become more common, allowing a greater standardisation of experimental approaches and genetic markers. GWA studies have been possible in the sheep, following the release of the Illumina Ovine 50 SNP BeadChip (50k SNP chip) in January 2009. This assay features over 54 000 probes that target single-nucleotide polymorphisms (SNPs) across the genome, providing genome-wide coverage with an estimated one marker per 46 kb on average (Illumina, 2010; http://www.illumina.com/products/ovine_snp50_whole_genome_genotyping_kits.ilmn).
The sheep 50k SNP chip has already been demonstrated as providing the ability to map causal mutations for traits showing simple patterns of inheritance (Becker et al., 2010). However, in sheep, as well as in other species including humans, answers have not been so definitive for complex traits and GWA studies have generally failed to explain most of the known genetic variation influencing complex diseases (for example, Manolio et al., 2009; Kemper et al., 2011). These studies typically test each marker independently for an association with the trait. The expectation is that the variance explained by each marker is proportional to the size of the effect of the (unobserved) polymorphism on the trait, the degree of association between the marker and the polymorphism, and the experimental error associated with the measurement. Attempts to increase the power of association studies have focused either on increasing the number of markers or the number of observations for a trait. Few have attempted to formally estimate the distribution of marker effects with dense SNP markers (for example, Hayes et al., 2010; Kemper et al., 2011) and thus the required power of experiments for complex disease traits, such as nematode resistance, is still unknown. An alternate approach exploiting dense SNP chip data, known as Regional Genomic Relationship Mapping or Regional Heritability Mapping (RHM) (Nagamine et al., 2012), has been advanced as a better approach to capture more of the underlying genetic effects. This method provides heritability estimates attributable to small genomic regions, and it has the power to detect regions containing multiple alleles that individually contribute too little variance to be detected by GWA studies.
The aim of this study was to identify loci underlying variation in FEC and IgA, as indicators of nematode resistance, and body weight in Scottish Blackface lambs, genotyped with the 50k SNP chip. The lambs are grouped in several large half-sib families within a complex pedigree, and thus provide a suitable population for using and comparing both the GWA and RHM approaches.
Materials and methods
Population
The population consisted of 752 Scottish Blackface lambs, bred over a 3-year period (2001–2003) with both sire and dam recorded at birth for all animals, and a complete pedigree available back to the foundation of the flock in 1988. Previously, this flock had been divergently selected for carcass lean content from 1988 to 1996, creating lean and fat lines, after which the lines were maintained as closed populations with no further selection (Bishop, 1993). The genotyped population comprised F2 and double backcross lambs from these lines, bred from 10 sires (half-sib family size=11–146) using natural mating. Lambs were born on pasture, over a 4-week period each year, and were continuously exposed to natural mixed nematode infection. Two weeks after commencement of lambing, the ewes who had yet to lamb were moved to a new pasture, creating two management groups. After weaning at ∼12 weeks of age, the lambs were run as one flock again. Husbandry procedures followed standard commercial practice.
Phenotypic measurements
Faecal samples were collected from the rectum of the lamb at approximately 16, 20 and 24 weeks of age (that is, in August, September and October). These samples were used for FEC assays, using the modified McMaster technique (Gordon and Whitlock, 1939; Bairden, 1991), which distinguishes parasite genera on the basis of egg size and morphology. FEC were measured on four replicates of each faecal sample and the average of the four replicates at each time point was used for analyses. Eggs were classified according to whether they were Nematodirus spp. or other nematode genera collectively termed Strongyles, the latter potentially including Oesophagostomum, Chabertia, Bunostomum, Trichostrongylus, Cooperia, Ostertagia, Teladorsagia, and Haemonchus genera. The egg counts for Nematodirus spp. were recorded separately, as the eggs have a distinct size and shape, and could be visually distinguished. At the time of sampling, body weight was recorded for all individuals.
For 737 out of the 752 animals, measurements on the activity of plasma IgA against a somatic extract of third-stage larvae from T. circumcincta were available. They were measured by indirect enzyme-linked immunosorbent assay, as described by Strain et al. (2002), on blood samples collected at 24 weeks of age. Relative IgA activity was measured as: (observed–standard)/(high control–standard), where the observed value is the sample mean from three replicates for the animal, the standard is the mean of three replicates from a pooled sample of helminth-naive lambs and the high control is the mean of three replicates from a pool of high-responder lambs (Sinski et al., 1995). The pool of high-responder lambs was created by combining equal quantities of plasma from six lambs that gave strong IgA responses following natural infection. The value for each animal was therefore expressed as a proportion of a positive control.
Genotype data
Animals were genotyped using the 50k SNP chip, and genotypes analysed with two different approaches, GWA and RHM, as described below. The SNP genotype data were subjected to quality control measures as follows. The quality control removed the SNPs with a minor-allele frequency <0.02 or a call rate <95%. Deviation from Hardy–Weinberg equilibrium was not considered as a method for excluding SNPs. Furthermore, markers on the sex chromosome were removed from the analysis. After quality control, 44 388 SNPs were available for the GWA analysis and 42 841 for the RHM. The difference in number was owing to the fact that the GWA analysis uses all SNPs, regardless of whether they were mapped or not, whereas RHM requires mapped SNPs. Positions of SNP markers were obtained from the Sheep Genome browser v2.0 (http://www.livestockgenomics.csiro.au/sheep/).
Statistical analyses
FEC and IgA measurements were all right-skewed. Thus, before analyses, FEC measurements were log-transformed to  , where x is a constant used to avoid zero values, and IgA data were cube-root transformed. Further, in addition to time-point-specific data, averaged data across the three time points were also analysed for FEC (both Nematodirus and Strongyles) and body weight. This ‘average animal effect’ for each trait was estimated by fitting a repeatability model to trait values across the three ages.
, where x is a constant used to avoid zero values, and IgA data were cube-root transformed. Further, in addition to time-point-specific data, averaged data across the three time points were also analysed for FEC (both Nematodirus and Strongyles) and body weight. This ‘average animal effect’ for each trait was estimated by fitting a repeatability model to trait values across the three ages.
Owing to the previous selection history of the flock, it was important to identify and correct for possible population stratification. Classical multi-dimension scaling was used to explore population substructure and to verify the genetic homogeneity of the sample before analysis. As this analysis did not reveal any obvious substructure, this aspect was not considered in further analyses, other than correct for kinship, using the genomic kinship matrix and correct P-values with the genomic inflation factor λ, as described below.
GWA analyses were performed using the GenABEL package (Aulchenko et al., 2007) in R environment (http://www.r-project.org). The first step consisted of fitting mixed models with both fixed and polygenic effects to each trait, the latter to account for genetic relationships between animals. The fixed effects were: sex (male and female), year (2001–2003), management group (two levels, viz. those born in the first 2 weeks of the lambing season and those born subsequently), litter size (single or multiple) and age of dam (1–4 years), with day of birth as covariate. To account for relatedness, the variance/covariance matrix was estimated from the genomic kinship matrix, constructed using pair-wise identities by state, calculated for all samples on the basis of all autosomal SNPs, as implemented in the GenABEL package (Aulchenko et al., 2007). Second, association was tested using an mmscore function (Chen and Abecasis, 2007) on the residuals, which have been corrected for familial relatedness, and thus should be independent of pedigree or prior selection. After Bonferroni correction, significance thresholds were P<1.13 × 10−6 and P<2.25 × 10−5 for genome-wide (P<0.05) and suggestive (that is, one false positive per genome scan) levels, corresponding to –log10(P) of 5.95 and 4.65, respectively. P-values were corrected for the genomic inflation factor λ, which accounts for any systematic deviations of observed vs expected P-values (possibly because of remaining population substructure not captured by the genomic kinship matrix). Because mmscore SNP effects are biased downwards, effects for a subset of significant SNPs were re-estimated in ASReml (Gilmour et al., 2009) by fitting the SNPs, one at a time, as fixed effects in the same model as used for the GWA analyses. Effects were calculated as follows: additive effect, a=(AA−BB)/2; dominance effect, d=AB−((AA+BB)/2); and proportion of VA due to each SNP was estimated as (2pq (a+d (q−p))2)/VA, where AA, BB and AB were the predicted trait values for each genotype class, p and q were the allelic frequencies at the SNP locus and VA was the total additive genetic variance of the trait obtained when no SNP fixed effects were included in the model.
The second approach used to analyse the genotype data was RHM, in which each chromosome (OAR) is divided into windows of a pre-defined number of SNPs, and the variance attributable to each window estimated. In our case, the window size was initially 100 adjacent SNPs, and the window was shifted every 50 SNPs, excluding those regions at the end of the chromosome with <25% of the target number of SNPs. A mixed model was used for the analysis, accounting for the same fixed effects as above, with residual and additive genetic (both regional genomic and whole genomic) effects fitted as random. The whole genomic additive effect was estimated using the genomic relationship matrix constructed from all SNPs, whereas the regional genomic additive effect was estimated from a genomic relationship matrix constructed from the SNPs within each window, that is, region. Whole genomic, regional genomic and residual variances were σ2a, σ2w and σ2e, respectively. Phenotypic variance, σ2p, was then given by σ2a+σ2w+σ2e. Whole-genomic heritability was estimated as h2a=σ2a/σ2p, whereas the regional heritability was h2w=σ2w/σ2p.
To test for the differences in regional variance in the RHM approach, a likelihood ratio test (LRT) was used to compare a model fitting variance in a specific window (fitting both whole-genome and region-specific additive variance) against the null hypothesis of no variance in that window (whole-genome additive variance only). The test statistic was assumed to follow a mixture of  and
and  distributions (Self and Liang, 1987). In total, 868 windows were tested, of which half were used in the Bonferroni correction, to account for the overlapping windows. Hence, after Bonferroni correction to account for multiple testing, the LRT thresholds were 13.56 and 9.29, corresponding to P-values of P<1.15 × 10−4 and P<2.30 × 10−3 (corresponding to −log10(P) of 3.94 and 2.64), for genome-wide and suggestive significance levels, respectively. For windows that were declared significant by the criteria described above, as well as those with a LRT just below the significance level, smaller windows (that is, 50 SNP wide windows shifted every 25 SNPs) were used to test for improved mapping resolution. In this case, the LRT thresholds for genome-wide and suggestive significance levels were 14.87 and 10.57, corresponding to P-values of P<5.76 × 10−5 and P<1.15 × 10−3 (corresponding to –log10(P) of 4.24 and 2.94), respectively.
distributions (Self and Liang, 1987). In total, 868 windows were tested, of which half were used in the Bonferroni correction, to account for the overlapping windows. Hence, after Bonferroni correction to account for multiple testing, the LRT thresholds were 13.56 and 9.29, corresponding to P-values of P<1.15 × 10−4 and P<2.30 × 10−3 (corresponding to −log10(P) of 3.94 and 2.64), for genome-wide and suggestive significance levels, respectively. For windows that were declared significant by the criteria described above, as well as those with a LRT just below the significance level, smaller windows (that is, 50 SNP wide windows shifted every 25 SNPs) were used to test for improved mapping resolution. In this case, the LRT thresholds for genome-wide and suggestive significance levels were 14.87 and 10.57, corresponding to P-values of P<5.76 × 10−5 and P<1.15 × 10−3 (corresponding to –log10(P) of 4.24 and 2.94), respectively.
In addition to the two approaches of estimating genetic variances using the genomic relationship matrix, a variance component analysis was carried out in ASReml (Gilmour et al., 2009), using the complete available pedigree, to allow comparison of different approaches of defining genetic relationships. The model used accounted for the same fixed effects as previously described, with animal fitted as random. Although the relatively small number of sire families is not optimal for pedigree-based heritability estimation, that is, the s.e. of the estimate will be larger than if the population comprised more families, the data set does benefit from a deep pedigree extending back to 1988. A random litter (common environment) effect was also fitted in the analysis for body weight traits.
Results
Summary statistics for FEC, IgA and weight traits are reported in Supplementary Table 1. As expected, Nematodirus egg counts were considerably lower than Strongyles egg counts. The log-transformation was more successful in removing skewness in the Strongyles than the Nematodirus FEC, with the high proportion of zero values for Nematodirus FEC making it difficult to find a suitable transformation.
Heritabilities estimated using both a pedigree-based approach and the genomic relationship matrix from the GenABEL and RHM analyses are reported in Supplementary Table 2. In general, the estimates obtained with the three methods were comparable, with a tendency for the pedigree-based estimates to be higher.
Genome-wide association analysis
GWA analysis identified three SNPs with genome-wide significant association, these being for Nematodirus average animal effect on OAR 14 (corrected P-value=6.03 × 10−7, −log10(P)=6.22), Strongyles FEC at 16 weeks on OAR 6 (corrected P-value=6.56 × 10−7, −log10(P)=6.18) and body weight average animal effect on OAR 21 (corrected P-value=5.06 × 10−7, −log10(P)=6.30). However, several SNPs reached the suggestive level, by the definitions we used. A summary of the identified SNPs, the trait they were associated with, their map locations and their P-values are reported in Table 1 for IgA and FEC traits and in Table 2 for body weight traits, tabulating P-values <9.95 × 10−5 (corresponding to −log10(P) of 4.00). Not surprisingly, some SNPs were found to be important in more than one trait (colour marked in Tables 1 and 2), that is, both within the same parasite species and within body weight, although the significance levels varied with trait.
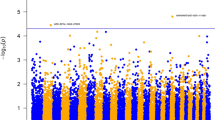
Manhattan plots displaying the GWA results and the corresponding Q–Q plots of observed P-values against expected P-values for Nematodirus average animal effect, Strongyles FEC at 16 weeks and body weight average animal effect are shown in Figures 1, 2, 3, respectively, as examples of results achieving genome-wide significance.
Manhattan plot displaying the GWA results (−log10(P) of the corresponding Pc1df, P-values corrected for the genomic inflation factor λ) and Q–Q plot (below) of observed P-values against the expected P-values for Nematodirus average animal effect. Genome-wide P<0.05 (solid line) and suggestive (dashed line) thresholds are also shown.
Manhattan plot displaying the GWA results (−log10(P) of the corresponding Pc1df, P-values corrected for the genomic inflation factor λ) and Q–Q plot (below) of observed P-values against the expected P-values for Strongyles FEC at 16 weeks. Genome-wide P<0.05 (solid line) and suggestive (dashed line) thresholds are also shown.
Manhattan plot displaying the GWA results (−log10(P) of the corresponding Pc1df, P-values corrected for the genomic inflation factor λ) and Q–Q plot (below) of observed P-values against the expected P-values for body weight average animal effect. Genome-wide P<0.05 (solid line) and suggestive (dashed line) thresholds are also shown.
Significance test results (P-values) from the SNPs identified in the GWA analysis and warranting further analysis in ASReml are given in Supplementary Table 3. As shown (in bold), significance levels were confirmed for all SNPs identified in the GWA analysis. However, it is interesting to note that SNPs significant for one trait were in general significance (although the significance levels varied with trait) for the other traits within the same parasite species and for body weight at different ages. In particular, five SNPs were specific for Strongyles, 13 for Nematodirus, three for IgA and four for body weight. However, there were cases where SNPs were also significant across trait categories, that is, eight SNPs were associated with both Strongyles and Nematodirus; five associated with Strongyles and body weight; and one SNP was associated with Strongyles and IgA. Further, three SNPs were associated with both Nematodirus and IgA and with Nematodirus and body weight; and one SNP with IgA and body weight. Of interest, three SNPs affected more than two traits. Genetic effects were estimated for SNPs significant at the genome-wide level (results not shown). These SNPs do not display significant evidence for dominance and the proportion of the observed genetic variation explained was 0.19, 0.16 and 0.08, for the SNPs associated with Nematodirus average animal effect, Strongyles FEC at 16 weeks and body weight average animal effect, respectively.
Regional heritability mapping analysis
The results for the RHM analysis using 100 SNP window size for nematode resistance and weight traits are given in Tables 3 and 4, respectively. A region on OAR 14 (window 14) was significant (LRT=20.54) at the genome-wide level (P<0.05) for Nematodirus (average animal effect), and seven other regions reached the suggestive level (Table 3). Moreover, a region on OAR 14 was also significant (LRT=20.46) at the genome-wide level for Nematodirus FEC at 24 weeks, when analysed with 50 SNP window size (results not shown). Regional heritabilities varied from 0.04 to 0.09. As an example, Figure 4 shows the LRT across the whole genome in Figure 4a, as well as a snapshot on OAR 14 (Figure 4b) for Nematodirus average animal effect.
Plot of the LRT across the genome (a) and for chromosome 14 (b) for Nematodirus average animal effect. The windows indicated by the purple rectangle in plot (b) were analysed using smaller windows (that is, 50 SNPs). These windows are reported in plot (c) and highlighted by a purple rectangle. Genome-wide P<0.05 (in green) and suggestive (in red) thresholds are also shown in (b) and (c). In plots (b) and (c), the first SNP identified by the purple rectangles is OAR14_40458094.1 (position 38703731 bp) and the last one is s26895.1 (position 51406211 bp).
Strongyles results revealed that three regions (window 13 on OAR 6 for 16 weeks FEC; window 33 on OAR 3 and window 14 on OAR 21 for the average animal effect) reached the suggestive level, but with no regions reaching the genome-wide significance level. For Strongyles FEC traits, the highest regional heritability (0.07) corresponded to the region (that is, window 14 on OAR 21) with LRT of 13.44. With no regions reaching either the suggestive or genome-wide significance levels for IgA, weak signals from the 100 SNP window analysis found on OAR 3 and 4 regions formed the basis for further analyses using smaller window size.
Results for body weight traits revealed two regions (12 and 13) on OAR 6 reaching the suggestive level (LRT of 9.58–12) at 16 weeks. These same regions (windows 12 and 13) had LRT just below the significance threshold for the other body weight traits; hence, we decided to explore them further with smaller windows in all body weight traits. In addition, window 14 on OAR 6 reached a LRT of 9.24 for 16 week body weight, as well as window 10 on OAR 3 for weight at 20 weeks. Two other windows, 32 on OAR 8 for body weight at 20 weeks and 12 on OAR 25 for the average animal effect, had LRT of 9.58 and 9.34, respectively. These two regions were those with the highest regional heritability for the body weight traits (that is, 0.07).
Apart from the result for Nematodirus FEC at 24 weeks, 50 SNP window-size analyses tended, in general, to confirm those from the larger window size. Regions that were at suggestive significance level with the 100 SNP window size were still significant with the 50 SNP window size. Moreover, the IgA regions were still not significant. However, from the two windows on OAR 14 that were at the suggestive (LRT=11.17) and genome-wide (LRT=20.54) significance level, respectively, for Nematodirus average animal effect, four windows became significant at the genome-wide level (with LRT range increasing to 18.78–31.13). In the latter case, these four windows included areas of the genome that were not significant before. As an example, Figure 4c shows the LRT for the 50 SNP window analysis, as an enlargement of three genome regions (indicated by a box in Figure 4b) identified with the 100 SNP window-size analysis.
Discussion
Both GWA and RHM approaches used in this study identified genomic regions of interest for nematode resistance and body weight traits in Scottish Blackface lambs, with both methods performing well when there was a strong evidence of association. For example, both methods identified similar regions for SNPs associated with Nematodirus average animal effect (OAR 14), Strongyles FEC at 16 weeks (OAR 6) and body weight at 16 weeks (OAR 6). However, there were some subtle differences between the two methods. For example, the GWA analysis identified a genome-wide significant SNP associated with a body weight trait and a SNP reaching the suggestive level for Nematodirus that were not detected by RHM. On the other hand, several exclusive regions reaching the suggestive level were identified only with RHM. Nagamine et al. (2012) showed that RHM performed better than a standard GWA study, especially when associated SNPs do not have large enough effect to be declared significant at the genome-wide level. In principle, estimating the trait heritability for chosen regions allows integration of the variance contributed by both rare and common variants into a single estimate of additive variance. Because, the RHM combines different sources of variation within the region, it potentially allows the identification of loci that cannot be found by GWA analysis. Therefore, RHM should have greater power than a standard GWA analysis to map genomic regions that contribute variance due to the segregation of several common or rare variants while retaining similar power to map loci segregating for a single common variant.
Similar genetic parameter estimates obtained from conventional pedigree-based analysis, and both GWA and RHM approaches, confirmed the power of using the genomic kinship matrix to quantify genetic variation in pedigree structured data. Although the pedigree-based estimates tended to be higher, this may be explained by the data structure. The pedigree-based method is heavily dependent on the number and properties of the sire families (that is, sire averages), and there were few sire families in our data set. On the other hand, the marker-based methods better capture within-family genetic variation, potentially giving unbiased estimates of genetic effects for each individual within a family.
Considerable work has been carried out to address the genetic control of resistance to nematodes, with the majority of studies being microsatellite-based linkage studies (for example, Dominik, 2005; Crawford et al., 2006; Davies et al., 2006). More recently, GWA studies using higher density SNP panels have become more common. To our knowledge, however, few studies have been published so far using SNP chip data in sheep (for example, Kemper et al., 2011; Sallé et al., 2012). Comparisons across studies are not easy, given the complexity of the trait and the differences among populations and approaches used. Moreover, in contrast to human association analysis where, in general, small well-defined families or unrelated individuals are used, the design of genome-wide scans in livestock is often based on field samples with inaccurate pedigree information, posing a series of challenges. Livestock populations are characterised by high levels of relatedness, that is, closely related animals with a complex population structure and an a priori unbalanced distribution of allele frequencies. This is even more challenging in this study, in which the number of sires used was small. This aspect is likely to inflate the rate of false-positive associations between the trait and the markers, making true associations harder to detect.
Some of the regions previously reported by Davies et al. (2006) for nematode resistance using a partial genome scan with microsatellite markers in a within-family linkage analysis were also confirmed with SNPs using both GWA and RHM approaches. This is despite the fact that linkage and linkage disequilibrium (LD)-based analysis methods combine genomic information in different ways, that is, utilising within-family transmission of regions linked to phenotypes vs population-wide LD between marker and causative mutation. For example, a SNP reaching the suggestive level for Nematodirus FEC at 20 weeks on OAR 2 was identified by the GWA approach, close to the QTL region reported by Davies et al. (2006) for the same trait and population.
One of our most significant results, on OAR 14 for Nematodirus average animal effect, was also detected by both methods. Of interest is to note that although a block of ∼8 SNPs (Figure 1) was detected with GWA analysis (one genome-wide significant SNP and two SNPs reaching the suggestive level), only a few of these SNPs show high pairwise linkage disequilibrium, that is, r2>0.3 (see Supplementary Table 4). This result was confirmed by RHM, which identified two regions, one significant at the genome-wide and the other reaching the suggestive level. Although the locations identified by the two methods are not completely overlapping, they fell in the marker region, reported by Davies et al. (2006), in the same population and, reported by Matika et al. (2011), in Texel and Suffolk sheep. Moreover, it is interesting to note that when using smaller windows (that is, 50 SNPs) with RHM, the region found to be significant is wider, including also the positions identified by the GWA analysis. This region has now been confirmed in three separate studies and represents one of the most convincing regions associated with resistance to nematode infection. Unfortunately, at present, there appear to be no obvious candidate genes within this region, using either the UCSC genome browser (http://genome.ucsc.edu/) or the CSIRO sheep genome browser (http://www.livestockgenomics.csiro.au/cgi-bin/gbrowse/oarv2.0/). Recently, however, in a comparative genomics study of toll-like receptor signalling in five species (human, mouse, pig, cattle and sheep), Jann et al. (2009) assigned interferon regulatory factor (IRF3) using both in-silico and radiation hybrid mapping to OAR14, within the region covered by our markers. These authors located IRF3 to a region affecting health traits in the five species considered, with host QTL controlling a wide range of pathogens, making it a compelling candidate gene for our OAR14 QTL.
To our knowledge, there are no other published QTL results for Nematodirus; most of the reported studies consider Strongyles genera, such as Trichostrongylus and Haemonchus. However, positive and moderate to strong genetic correlations between Nematodirus and Strongyles FEC ranging from 0.49 to 0.93 have been reported (Bishop et al., 2004, Morris et al., 2004, Wolf et al., 2008). This implies, therefore, some common genetic mechanisms responsible for resistance to Strongyles and Nematodirus; hence, it may be hypothesised that some SNPs or QTL underlying genetic variation in Nematodirus and Strongyles FEC may be in common. In the current study, no SNPs in common for both Nematodirus and Strongyles were identified by GWA and RHM approaches. However, some SNPs analysed using a full mixed model analysis in ASReml showed significance across categories, albeit at relaxed significance criteria.
As was the case with Nematodirus, both GWA and RHM approaches identified similar significant QTL regions for Strongyles. For example, a genome-wide significant SNP for Strongyles FEC at 16 weeks (on OAR 6) detected by GWA analysis was also confirmed by RHM with the latter also identifying two more suggestive regions for Strongyles average animal effect (OAR 3 and 21). The region on OAR 6 is close to the secreted phosphoprotein 1 (SPP1) gene, which is known to be involved in the immune response (Li et al., 2011). Regions on OAR 6 have been previously reported by Beh et al. (2002) and Beraldi et al. (2007), above and below the suggestive threshold, respectively. Moreover, these authors have identified regions on OAR 1 and 12, although these fell below the suggestive threshold. However, strong evidence for a region on OAR 12 affecting FEC was recently reported by Sallé et al. (2012); in a study investigating experimentally infected Martinique Black Belly × Romane sheep, a cluster of highly significant SNPs on OAR 12 was associated with FEC. However, in the current study, no significant SNPs for either Nematodirus or Strongyles FEC on OAR 12 were detected with either approach. In addition, Sallé et al. (2012) reported a QTL on OAR 3 associated with worm burden and FEC around 150 Mbp. We also identified a SNP in the same region for Nematodirus FEC at 24 weeks, which was below the significance threshold, but was highly significant (P<0.0001) when further tested as a single SNP in ASReml. Moreover, we identified two SNPs (although below the suggestive significance level) on OAR 3 associated with IgA, which were also significant for FEC traits when analysed in ASReml. Although no causative mutation has been identified for these OAR3 QTL, the effect is often presumed to be mediated through INFG in sheep (Stear et al., 2009).
Although, the major histocompatibility complex, on OAR 20, has often been proposed as a functional candidate for nematode resistance (for example, Schwaiger et al., 1995; Buitkamp et al., 1996), the traits we considered were not associated with the major histocompatibility complex at genome-wide or at suggestive level in our study. The only SNP detected on OAR 20 with a suggestive significance threshold for Strongyles FEC at 16 weeks was not close to the major histocompatibility complex region.
Several QTL for body size or growth rate have been reported in livestock, with the majority of the studies using univariate approaches, treating weights recorded at a particular growth point as separate traits. In sheep, results have been reported for a wide variety of breeds and crosses, approaches (GWA vs linkage) and trait definitions (for example, Walling et al., 2004; McRae et al., 2005; Karamichou et al., 2006; Raadsma et al., 2009; Jonas et al., 2010). However, Hadjipavlou and Bishop (2008) have shown that body weight QTL changes over time in Scottish Blackface sheep. In particular, these authors reported QTL on OAR 3 and 14 with significant effects during early growth, whereas a QTL on OAR 20 had significant effects on growth variables around intermediate/maximum growth. The region identified on OAR 14 corresponds to the region identified by Davies et al. (2006) for Nematodirus FEC, which is therefore close to the region we identified. We did not identify any SNP associated with body weight traits with either method in this region. However, one SNP on OAR 14, that was below the suggestive significance threshold for Nematodirus, showed significance for both Nematodirus and body weight, when tested using full mixed models in ASReml.
Although, no in-depth studies were conducted on pleiotropic relationships between traits, a region on OAR 6 associated with Strongyles FEC at 16 weeks was also associated with body weight traits. This result is consistent with the presence of non-zero genetic correlations between body-weight and indicator resistance traits reported in several field studies (for example, Bisset et al., 1992; Bishop et al., 1996). However, it should be noted that the SNP associations are only capturing a proportion of the observed trait genetic variation; therefore, even though genetic correlations suggest the existence of pleiotropic loci, such loci may well be missed in analyses such as these.
In summary, this study has been successful in detecting QTL for nematode resistance and body weight, using two contrasting methods of analysing SNP chip data. However, as in most GWA studies, there is a ‘missing heritability’ component, that is, the experiment only has the power to detect and declare significant a small proportion of the genetic variation. Further, there is some inconsistency of results between this and other published experiments. Partly this may be because of the differences in experimental design and the genetic background of different populations; however, it is also likely to be due, in part, to the complexity of the traits: because FEC is the net outcome of many physiological and immunological pathways, we may expect genetic differences between animals to be owing to variation at a large number of genes. Commonality of genetic mechanisms may perhaps be best explored using meta-analyses of independent data sets. This study is part of a larger European project aiming at identifying SNPs/regions in several independent populations and breeds. These data sets should afford the opportunity for such a meta-analysis, which, if successful, may even identify loci that could be directly used for breeding purposes.
Empirical evidence of complexity of genetic control of FEC and live weight may also be inferred from the RHM results. Nagamine et al. (2012) suggested that the use of smaller windows can improve mapping resolution. In our study, this was not always the case; that is, when moving from 100 to 50 SNP wide windows, only in a few cases, there was an improvement in the level of significance—sometimes regions became not significant. This is presumably a function of the architecture of the trait, that is, the number and size of SNP effects, the allele frequencies and the patterns of LD between the causative mutations and the SNPs. Failure to increase significance when moving to slightly narrower windows is suggestive of polygenic inheritance, that is, the observed effects being due to a large number of loci, each with a small effect. This conclusion is in agreement with that of Kemper et al. (2011).
Data archiving
Data are deposited in the Dryad repository: doi:10.5061/dryad.8f191.
References
Aulchenko YS, Ripke S, Isaacs A, van Duijn CM (2007). GenABEL: an R library for genome-wide association analysis. Bioinformatics 23: 1294–1296.
Bairden K (1991). Ruminant parasitic gastroenteritis: some observations on epidemiology and control. PhD Thesis, University of Glasgow.
Bartley DJ, Jackson F, Jackson E, Sargison N (2004). Characterization of two triple resistant field isolates of Teladorsagia from Scottish lowland sheep farms. Vet Parasitol 123: 189–199.
Becker D, Tetens J, Brunner A, Burstel D, Ganter M, Kijas J et al (2010). Microphthalmia in Texel sheep is associated with a missense mutation in the paired-like homeodomain 3 (PITX3) gene. PLoS One 5: e8689.
Beh KJ, Hulme DJ, Callaghan MJ, Leish Z, Lenane I, Windon RG et al (2002). A genome scan for quantitative trait loci affecting resistance to Trichostrongylus colubriformis in sheep. Anim Genet 93: 97–106.
Beraldi D, McRae AF, Gratten J, Pilkington JG, Slate J, Visscher PM et al (2007). Quantitative trait loci (QTL) mapping of resistance to strongyles and coccidia in the free-living Soay sheep (Ovis aries). Int J Parasitol 37: 121–129.
Besier RB, Love SCJ (2003). Anthelmintic resistance in sheep nematodes in Australia: the need for new approaches. Aust J Exp Agr 43: 1383–1391.
Bishop SC (1993). Selection for predicted carcass lean content in Scottish Blackfae sheep. Anim Prod 56: 379–386.
Bishop SC, Bairden K, McKellar QA, Park M, Stear MJ (1996). Genetic parameters for faecal egg count following mixed, natural, predominantly Ostertagtia circumcincta infection and relationships with live weight in young lambs. Anim Sci 63: 423–428.
Bishop SC, Jackson F, Coop RL, Stear MJ (2004). Genetic parameters for resistance to nematode infections in Texel lambs and their utility in breeding programmes. Anim Sci 78: 185–194.
Bishop SC, Stear MJ (2001). Inheritance of, and factors affecting, egg counts during early lactation in Scottish Blackface ewes facing mixed, natural nematode infections. Anim Sci 73: 389–395.
Bisset SA, Vlassof A, Morris CA, Sourthey BR, Baker RL, Parker AGH (1992). Heritability of and genetic correlations among faecal egg counts and productivity traits in Romney sheep. New Zeal J Agric Res 35: 51–58.
Buitkamp JFP, Stear MJ, Epplen JT (1996). Class I and class II major histocompatibility complex alleles are associated with faecal egg counts following natural, predominantly Ostertagia circumcinca infection. Parasitol Res 82: 693–696.
Chen WM, Abecasis GR (2007). Family-based association tests for genome-wide association scans. Am J Hum Genet 81: 913–926.
Coltman DW, Wilson K, Pilkington JG, Stear MJ, Pemberton JM (2001). A microsatellite polymorphism in the gamma interferon gene is associated with resistance to gastrointestinal nematodes in a naturally-parasitized population of Soay sheep. Parasitology 122: 571–582.
Crawford AM, Paterson KA, Dodds KG, Diez-Tascon C, Williamson PA, Roberts Thomson M et al (2006). Discovery of quantitative trait loci for resistance to parasitic nematode infection in sheep: I. Analysis of outcross pedigrees. BMC Genomics 7: 178.
Davies G, Stear MJ, Benothman M, Abuagob O, Kerr A, Mitchell S et al (2006). Quantitative trait loci associated with parasitic infection in Scottish blackface sheep. Heredity 96: 252–258.
Diez-Tascon C, Macdonald PA, Dodds KG, McEwan JC, Crawford AM (2002). A screen of chromosome 1 for QTL affecting nematode resistance in an ovine outcross population. Proc 7th World Congr Genet Appl Livest Prod Communication 13–37.
Dominik S (2005). Quantitative trait loci for internal nematode resistance in sheep: a review. Genet Sel Evol 37: S83–S96.
Gilmour AR, Gogel BJ, Cullis BR, Thompson R (2009) ASReml User Guide Release 3.0 VSN International Ltd, Hempstead, HP1 1ES, UK.
Goddard ME, Hayes BJ (2009). Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat Rev Genet 10: 381–391.
Gordon HM, Whitlock HV (1939). A new technique for counting nematode eggs in sheep faeces. J Council Sci Ind Res Australia 12: 50.
Hadjipavlou G, Bishop SC (2008). Age-dependent quantitative trait loci affecting growth traits in Scottish Blackface sheep. Anim Genet 40: 165–175.
Hayes BJ, Pryce J, Chamberlain AJ, Bowman PJ, Goddard ME (2010). Genetic architecture of complex traits and accuracy of genomic prediction: coat colour, milk fat percentage, and type in Holstein cattle as contrasting model traits. Plos Genet 6: e1001139.
Illumina (2010) OvineSNP50 DNA analysis kits. Viewed 28th February 2010.
Jann OC, King A, Corrales NL, Anderson SI, Jensen K, Ait-Ali T et al (2009). Comparative genomics of Toll-like receptor signalling in five species. BMC Genom 10: 216.
Jonas E, Thomson PC, Raadsma HW (2010). Genome-wide association study and fine mapping of QTL on OAR 21 for body weight in sheep. Proceedings of the 9th World Congress on the Genetic Applications of Livestock Production Comm 0938.
Karamichou E, Richardson RI, Nute GR, McLean KA, Bishop SC (2006). A partial genome scan to map quantitative trait loci for carcass composition, as assessed by X-ray computer tomography, and meat quality traits in Scottish Blackface sheep. Anim Sci 82: 301–309.
Kemper KE, Palmer DG, Liu SM, Greeff JC, Bishop SC, Karlsson JLE (2010). Reduction of faecal worm egg count, worm numbers and worm fecundity in sheep selected for worm resistance following artificial infection with Teladorsagia circumcincta and Trichostrongylus colubriformis. Vet Parasitol 171: 238–246.
Kemper KE, Emery DL, Bishop SC, Oddy H, Hayes BJ, Dominik S et al (2011). The distribution of SNP marker effects for faecal worm egg count in sheep, and the feasibility of using these markers to predict genetic merit for resistance to worm infections. Genet Res 93: 203–219.
Li RW, Li C, Gasbarre LC (2011). The vitamin D receptor and inducible nitric oxide synthase associated pathways in acquired resistance to Cooperia oncophora infection in cattle. Vet Res 42: 48.
Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ et al (2009). Finding the missing heritability of complex diseases. Nature 461: 747–753.
Matika O, Pong-Wong R, Woolliams JA, Bishop SC (2011). Confirmation of two quantitative trait loci regions for nematode resistance in commercial British terminal sire breeds. Animal 5: 1149–1156.
McRae AF, Bishop SC, Walling GA, Wilson AD, Visscher PM (2005). Mapping of multiple quantitative trait loci for growth and carcass traits in a complex commercial sheep pedigree. Anim Sci 80: 135–141.
Morris CA, Bisset SA, Vlassoff A, West CJ, Wheeler M (1998). Faecal nematode egg counts in lactating ewes from Romney flocks selectively bred for divergence in lamb faecal egg count. Anim Sci 67: 283–288.
Morris CA, Bisset SA, Vlassoff A, West CJ, Wheeler M (2004). Genetic parameters for Nematodirus spp. egg counts in Romney lambs in New Zealand. Anim Sci 79: 33–39.
Nagamine Y, Pong-Wong R, Navarro P, Vitart V, Hayward C, Rudan I et al (2012). Localising loci underlying complex trait variation using regional genomic relationship mapping. PLoS ONE 7: e46501.
Nieuwhof GJ, Bishop SC (2005). Costs of the major endemic diseases of sheep in Great Britain and the potential benefits of reduction in disease impact. Anim Sci 81: 23–29.
Raadsma HW, Thomson PC, Zenger KR, Cavanagh C, Lam MK, Jonas E et al (2009). Mapping quantitative trait loci (QTL) in sheep. I. A new male framework linkage map and QTL for growth rate and body weight. Genet Sel Evol 41: 34.
Sallé G, Jacquiet P, Gruner L, Cortet J, Sauvé C, Prévot F et al (2012). A genome scan for QTL affecting resistance to Haemonchus contortus in sheep. J Anim Sci 90: 4690–4705.
Schwaiger FW, Gostomski D, Stear MJ, Duncan JL, McKellar QA, Epplen JT et al (1995). An ovine major histocompatibility complex DRB1 allele is associated with low faecal egg counts following natural, predominantly Ostertagia circumcincta infection. J Parasitol 25: 815–822.
Self SG, Liang KY (1987). Asymptotic properties of maximum-likelihood estimators and likelihood ratio tests under non standard conditions. J Amer Stat Assoc 82: 605–610.
Sinski E, Bairden K, Duncan JL, Eisler MC, Holmes PH, McKellar QA et al (1995). Local and plasma antibodyresponses to the parasitic larval stages of the abomasal nematode Ostertagia circumcincta. Vet Parasitol 59: 107–118.
Stear MJ, Boag B, Cattadori I, Murphy L (2009). Genetic variation in resistance to mixed, predominantly Teladorsagia circumcincta nematode infections of sheep: from heritabilities to gene identification. Parasite Immunol 31: 274–282.
Strain S, Bishop SC, Henderson NG, Holmes PH, McKellar QA, Mitchell S et al (2002). The genetic control of IgA activity against Teladorsagia circumcincta and its association with parasite resistance in naturally infected sheep. Parasitology 124: 545–552.
Walling GA, Visscher PM, Wilson AD, McTeir BL, Simm G, Bishop SC (2004). Mapping of quantitative trait loci for growth and carcass traits in commercial sheep populations. J Anim Sci 82: 2234–2245.
Wolf BT, Howells K, Nakielny C, Haresign W, Lewis RM, Davies O et al (2008). Genetic parameters for Strongyle and Nematodirus faecal egg counts in lambs and their relationships with performance traits. Livest Sci 113: 209–217.
Acknowledgements
We acknowledge funding from the 3SR project (Sustainable Solutions for Small Ruminants) – 7th Framework Programme. Call identifier FP7-KBBE-2009-3, Project 245140.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Heredity website
Rights and permissions
About this article
Cite this article
Riggio, V., Matika, O., Pong-Wong, R. et al. Genome-wide association and regional heritability mapping to identify loci underlying variation in nematode resistance and body weight in Scottish Blackface lambs. Heredity 110, 420–429 (2013). https://doi.org/10.1038/hdy.2012.90
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2012.90
Keywords
This article is cited by
-
Ovine PAPPA2 gene coding variants are linked to decreased fecal egg shedding in native Turkish sheep naturally infected with gastrointestinal nematodes
Tropical Animal Health and Production (2023)
-
Interaction networks and pathway analysis of genetic resistance to gastrointestinal nematodes in sheep
Tropical Animal Health and Production (2023)
-
Estimates of genomic heritability and genome-wide association studies for blood parameters in Akkaraman sheep
Scientific Reports (2022)
-
Genome-wide association study and genomic heritabilities for blood protein levels in Lori-Bakhtiari sheep
Scientific Reports (2021)
-
Immunoglobulins as Biomarkers for Gastrointestinal Nematodes Resistance in Small Ruminants: A systematic review
Scientific Reports (2020)