Abstract
Purpose: To evaluate genotype-phenotype correlation over time for a cohort of children with connexin-26 (GJB2)–associated autosomal recessive hearing loss.
Methods: Fifty-two children were identified from a database of individuals with homozygous or compound heterozygous mutations in GJB2 and subjected to chart review of their otolaryngologic and serial audiometric evaluations. Genotype-phenotype correlations were identified among the members of this group by appropriate statistical analyses.
Results: Hearing loss was most severe in individuals with two truncating mutations in GJB2 and mildest in those with two nontruncating mutations. Progressive hearing loss was seen directly by serial audiometry in 24% of all subjects, and suggested in a total of 28% when those with normal newborn hearing screens and subsequent hearing loss were included. Progression was particularly common among carriers of the p.V37I allele either in homozygosity or in compound heterozygosity with a truncating allele; these children are primarily of Asian descent and demonstrate mild, slowly progressive hearing loss.
Conclusions: Phenotype in GJB2-associated hearing loss is correlated with genotype, with truncating mutations giving rise to more severe hearing loss. Progression of hearing loss is not uncommon, especially in association with the p.V37I allele. These results suggest that close audiometric follow-up is warranted for patients with GJB2-associated recessive hearing loss.
Similar content being viewed by others
Main
Hearing loss is the most common congenital sensory deficit, affecting up to 1 in 300 newborns.1,2 A wide variety of environmental and genetic causes underlie congenital hearing loss, with familial deafness seen in both syndromic and nonsyndromic forms. Among >100 known forms of nonsyndromic deafness with identified genetic loci, by far the most common and best characterized is the one associated with GJB2 (MIM *121011), the gene encoding the connexin 26 protein.3,4 This gap junction protein, which assembles to form channels between cells, is thought to play a crucial role in K+ homeostasis and intercellular signaling within the organ of Corti.5 Almost 100 unique mutations in the GJB2 gene, which is located on Chromosome 13 of the human genome, are associated with recessive deafness. An additional 17 show a dominant inheritance pattern6 and often manifest as syndromic deafness, such as that seen in the keratitis-ichthyosis-deafness syndrome.7 The majority of GJB2-associated deafness, however, is nonsyndromic and exhibits an autosomal recessive inheritance pattern.
The best characterized of all GJB2 mutations is c.35delG, with a carrier rate of 2% to 4% in white individuals.8,9 Those who are homozygous for this mutation, which causes the premature truncation and effective absence of the connexin 26 protein, are typically affected with nonprogressive, severe-to-profound bilateral sensorineural hearing loss (SNHL) from birth. Because of its overwhelming abundance among Western people and its early description, the phenotype associated with the c.35delG mutation has served as the paradigm for GJB2-associated SNHL and guided its clinical management. In recent years, however, there has been increased recognition of the diversity of phenotypes associated with the various GJB2 genotypes. In particular, distinctions have been made between GJB2 mutant alleles based on their functional effect; that is, whether they constitute nonsense or missense mutations. Changes of the former type are truncating (T) mutations arising from the creation of a premature stop codon and result in the absence of any functional connexin 26 protein. Nontruncating (NT) mutations result in altered proteins whose functions may or may not be impaired.10 Comparison of >2000 patients in two large series showed significant differences in hearing thresholds between genotypes consisting of two truncating mutations (T/T), which were most severely impaired, those with two nontruncating mutations (NT/NT), which were least affected, and those with one of each (NT/T), which had an intermediate phenotype.10,11 In addition to this wide range of hearing impairment, there have been some reports of progressive hearing loss among patients with GJB2 mutations, despite the common notion that it is a nonprogressive condition.11,12
In this study, we present our experience with GJB2-associated hearing loss during 5 years at Lucille Packard Children's Hospital. Our pediatric patient population encompasses a wide range of ethnic backgrounds and thus a variety of GJB2 genotypes, as identified by DNA sequencing of the GJB2 gene. Complete otolaryngologic and audiometric analyses have permitted the evaluation of 52 individuals with biallelic mutations in GJB2. Our data confirm the genotype-phenotype correlations noted previously and provide additional evidence for progressive hearing loss, especially among those harboring the p.V37I allele. The diversity of phenotypes and propensity for progression observed in these patients advocate the close observation of all affected individuals and also highlight the clinical utility of obtaining genetic testing to help counsel parents and to inform long-term audiologic follow-up.
METHODS
Study population
Subjects were selected initially by compiling all homozygous and compound heterozygous GJB2 mutations identified by comprehensive sequencing of GJB2 Exon 2 performed by the Molecular Pathology Laboratory at Stanford University Medical Center during a 5-year period (2004–2009). Of these individuals, those with electronic medical records available documenting visits to the Audiology and Pediatric Otolaryngology clinics at Lucile Packard Children's Hospital were selected for analysis. From these records, we obtained demographic information, including ethnicity as determined by patient report, as well as all of the audiometric and clinical data presented in this study. For most analyses, including all audiograms and GJB2 sequencing results, primary data were used exclusively; newborn hearing screen (NHS) results were occasionally obtained by parental report. This study was approved by the Institutional Review Board of Stanford University.
Audiometric analysis
Unless otherwise specified, hearing thresholds from behavioral audiograms are reported as averages of pure-tone thresholds at 0.5, 1, 2, and 4 kHz in both ears (pure-tone average [PTA0.5–4]), a common audiologic measure that assesses hearing across speech frequencies. In cases where no responses were elicited due to the severity of the hearing loss, thresholds were approximated in the quantitative analysis as either the highest sound level tested, if reported, or the highest sound level produced by the apparatus (110 dB). This approximation was necessary for 14 patients with profound hearing loss and would only serve to underestimate the PTA0.5–4. In few instances, only auditory brainstem response (ABR) thresholds were available. For these, average thresholds in response to tone pips at 0.5, 1, 2, and 4 kHz were used in place of the PTA0.5–4. Throughout the study, only audiograms that had complete data for 0.5, 1, 2, and 4 kHz were included. Severity of hearing impairment was determined based on the PTA0.5–4 as follows: 21 dB to 40 dB was defined as “mild,” 41dB to 70 dB as “moderate,” 71 dB to 95 dB as “severe,” and >95 dB as “profound.”10
For serial audiometric analysis of progressive hearing loss, the averages of pure-tone thresholds at 0.5, 1, 2, 4, and 8 kHz (PTA0.5–8) from the earliest and most recent behavioral audiograms were compared. The 8-kHz pure-tone threshold was included in this analysis to increase the sensitivity of detection of progression at high frequencies. A few patients did not have data at the 8 kHz point; for these, complete datasets for PTA0.5–4 were used for longitudinal comparison. Comparisons for progression purposes were only made between audiograms with matching datasets.
The majority of patients received continuous audiologic care at a single facility. The test-retest variability in the PTA0.5–8 between audiograms was calculated for our study population as the standard deviation of successive intrasubject PTA0.5–8 thresholds for all nonprogressors. We chose to define progression as twice the test-retest variability in the PTA0.5–8 thresholds. By the central limit theorem, 95% of patients would be expected to lie within this range; those outside it were thus identified as significant outliers with progressive hearing loss. Rather than an arbitrary cutoff threshold shift, as has been used previously, this definition accounts specifically for our dataset and reliability of our audiometric testing. Those identified as progressors did, however, meet other previously reported thresholds for progression, including an absolute increase of 5 dB13 or of 1 dB/year.14 When possible, ABR and behavioral thresholds were only compared with like tests. For several patients, earlier ABRs were compared with later behavioral audiograms. Because ABRs tend to be biased toward higher thresholds, this comparison would only underestimate the degree of progression; however, none of the patients for whom the analysis compared ABR with behavioral thresholds were found to have progressive hearing loss.
Statistical analyses
Data are presented as means and 95% confidence intervals (CI). For analyses of nonparametric numeric values compared between two groups, the Mann-Whitney U rank-sum test was used; for comparison of more than two groups, the Kruskal-Wallis test was used; results are reported with both H values and the appropriate P value from the χ2 test. For categorical data, the Fisher exact test was used.
RESULTS
Demographics
During a 4-year period, we identified 73 children across a wide range of ethnic backgrounds with a mean age of 5.3 years at the time of genetic diagnosis (Fig. 1). All patients harbored either homozygous or compound heterozygous mutations in GJB2 except for three: one subject had one mutation each in GJB2 and GJB6, a combination known to cause hearing loss15,16; a second had one known pathologic mutation (p.V37I) along with a pair of polymorphisms in cis (p.V27I and p.E114G), which together are thought to act as a pathologic variant17; and a third was identified with a single autosomal dominant mutation associated with keratitis-ichthyosis-deafness syndrome (p.D50N).18 Of these 73, 18 were excluded from further analysis because the sequence changes identified (p.V27I, p.V153I, p.E114G, and p.I203T) are generally considered to be benign polymorphisms,6 and the autosomal dominant individual was excluded, as the associated phenotype is distinct from that typical for the autosomal recessive GJB2 mutations.7 Two patients had no audiologic evaluation available, leaving a total of 52 children with GJB2 mutations known to be associated with SNHL. These 52 children included four sibling pairs: two pairs of c.35delG homozygotes and one pair each of p.V37I/p.V37I and p.V37I/c.35delG. Because of the significant intrafamilial variability of connexin-26–associated hearing loss19 and the even distribution of the siblings across genotype classes, we opted to include all siblings in our analysis. Forty patients had documented computed tomography and/or magnetic resonance imaging, with only one abnormality noted—an enlarged vestibular aqueduct. This individual had symmetric hearing loss that was not thought to be due to the unilateral anatomic anomaly. Two other children had associated congenital abnormalities—isolated cleft lip and cleft palate in both cases. The ethnic makeup of the study group reflects the diversity of the general population of the San Francisco bay area—30% white, 45% Asian, and 23% Latino. Thirty-three of the 52 children had serial audiograms >4 months apart and were analyzed for progression of hearing loss.
Patient demographics. A, Flowchart of patient inclusion. Fifty-two patients had autosomal recessive disease-causing mutations in GJB2; of these, 33 had serial audiometric data. B, Demographics. Comorbidities identified: keratitis-ichthyosis-deafness syndrome, cleft lip/palate, and renal disease. Computed tomography (CT) abnormalities: enlarged vestibular aqueduct and asymmetry of the internal auditory canal.
Alleles and genotypes
The 52 patients represent a total of 24 alleles segregated into 21 genotypes (Fig. 2). Of the 24 alleles, 11 cause premature truncations (T) of the GJB2 gene product and 12 are nontruncating (NT) missense mutations. The final allele, del(GJB6-D13S1830), a large deletion in the adjacent GJB6 gene at the DFNB1 locus, was classified as a truncating mutation, as was done previously.10 The ethnic distribution of alleles conforms to previously described patterns, with c.35delG found predominantly in whites and p.V37I common among Asians. The genotypes comprise 5 with 2 truncating alleles (T/T; N = 14), 4 with 2 nontruncating mutations (NT/NT; N = 21), and 12 with 1 of each (NT/T; N = 17). These three subgroups, which have been shown previously to be phenotypically distinct,10 were designated for further analysis.
Allele and genotype. A, Distribution of alleles among the study population. NT, nontruncating; T, truncating. B, Genotypes represented among the 52 patients studied. T/T, truncating/truncating; NT/T, nontruncating/truncating; and NT/NT, nontruncating/nontruncating. C, Ethnic distribution of p.V37I and c.35delG alleles, the most common encountered in our study population. D, Ethnic distribution of genotypes.
Audiometric characteristics
The audiometric characteristics of these three groups are summarized in Figure 3. T/T individuals had the most severe hearing impairment, with a mean PTA0.5–4 threshold of 100.3 dB (95% CI: 92.4–108.2 dB). This was significantly different from both the NT/NT cohort, which had only mild hearing loss (26.5 dB; 95% CI: 22.7–30.4 dB), and the intermediate NT/T population (53.9 dB; 95% CI: 38.8–68.9 dB; H = 31.23; P < 0.001). Comparison of speech reception threshold (SRT) yielded similar results (T/T: 80.4, 95% CI: 70.1–90.7 dB; NT/T: 42.7 dB, 95% CI: 30.6–54.9 dB; NT/NT: 17.5 dB, 95% CI: 14.65–20.35 dB; H = 22.5; P < 0.001). Among the specific genotypes, individuals homozygous for c.35delG had consistently severe-to-profound hearing loss (N = 9; 104.3 dB; 95% CI: 99.4–109.2 dB), whereas those homozygous for p.V37I were relatively mildly impaired (N = 18; 26.3 dB; and 95% CI: 22.0–30.6 dB). There was a marked ethnicity-associated difference in GJB2-associated hearing loss, with whites (83.3 dB; 95% CI: 65.3–101.3 dB) and Latinos (82.2 dB; 95% CI: 66.8–99.7 dB) being much more severely affected than Asians (26.5 dB; 95% CI: 23.1–30.0 dB). These differences are not independent of genotype, but rather are due to ethnicity-associated differences in allele frequency, specifically the preponderance of the mild p.V37I allele among Asians and the high frequency of the severe c.35delG allele among whites.
The treatments pursued in these patients conformed to the degree of hearing loss and, thus, to genotype, even though there was no independent association of treatment with genotype. Patients in the most severely affected cohort, T/T, have all either undergone cochlear implantation (8 of 14; 57%) or received hearing aids (6 of 14; 43%). Even with their modest PTA threshold elevations, the majority of individuals in the NT/NT group required hearing aids (14 of 21; 67%), with only 33% (7 of 21) avoiding intervention entirely. The NT/T group was again intermediate, with 59% (10 of 17) using hearing aids, 17% (3 of 17) undergoing cochlear implantation, and 24% (4 of 17) not treated, reflecting the heterogeneity of this population.
Progression of hearing loss
The acquisition of serial audiometric data permitted the longitudinal analysis of hearing thresholds in 33 patients with an average interval of 21 months (range: 4–89 months; Fig. 4). Taken as a single group, there was no significant progression, with an average change in PTA0.5–8 threshold of 1.9 dB (95% CI: −0.1–3.9 dB) over the tested interval. When individual subjects were analyzed, however, there were eight children (24% of all subjects) with clear evidence of progression on serial audiometric analysis. All of these children had threshold shifts greater than twice the test-retest reliability of ±2.6 dB calculated directly from our audiologic data. All children had continuous audiologic care at a single facility, except one. This child had consistent progression noted at three facilities over 6 years. None of the eight progressors were related, and no abnormalities on computed tomography were noted among them. There was no significant difference in age at first audiogram, age of genetic diagnosis, or length of follow-up between the progression and nonprogression groups (Table 1).
Progressive hearing loss. A, Progressors (black) and nonprogressors (white) among the genotype groups and individuals with or without the p.V37I allele. B, Composite audiograms for the seven p.V37I-associated individuals found to have progressive hearing loss. Dashed line indicates audiogram at initial presentation. Solid line indicates interval audiogram demonstrating elevation of hearing threshold. C, NHS reports for subjects with NHS results available. White indicates PASS in at least one ear. Black indicates REFER in both ears. NHS, newborn hearing screening. D, Progressors (black) and nonprogressors (white) among individuals of each genotype group, as identified either by a serial audiogram or by a passed NHS, and subsequent hearing loss > 40 dB. *P < 0.05 by Fisher exact test.
The progressors included five individuals who were homozygous for p.V37I, with two nontruncating (NT/NT) mutations; one homozygous for c.35delG, a member of the T/T group; and two with one nontruncating and one truncating mutation: c.35delG/p.V37I and c.299_300delAT/p.V37I. This cohort of individuals with progressive hearing loss, who were followed up for an average of 29 months (range: 5–62 months), had a mean increase in PTA0.5–8 of 9.0 dB (95% CI: 6.8–11.1 dB). By comparison, the remaining patients exhibited no change in PTA0.5–8 threshold (−0.8 dB; 95% CI: −2.2–0.7 dB); these values are significantly different (P < 0.0001, Mann-Whitney U test). SRT changes were similar; the group of eight progressors had, on an average, 6.1 dB worsening in SRT (95% CI: 2.0–10.3 dB), compared with −0.4 dB (95% CI: −2.9–2.2 dB) for the nonprogressing group (P < 0.05, Mann-Whitney U test).
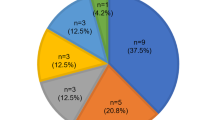
Although there was no significant difference in the overall PTA0.5–8 changes among the three main study groups, progression was more common among patients in whom the only nontruncated connexin 26 protein present harbors the p.V37I mutation; that is, those with either the p.V37I/p.V37I homozygous genotype or one with p.V37I together with a truncating allele (Fig. 4A). Of this group of 18 individuals, seven (39%) had progressive hearing loss, a significantly greater proportion than those with all other genotypes (1 of 15, 7%; P < 0.05). These seven individuals had a uniform phenotype; they all have mild hearing loss (37.5 dB; 95% CI: 30.7–44.4 dB) and a small but significant amount of progression (8.7 dB; 95% CI: 6.3–11.0 dB; Fig. 5). Five of the seven (72%) required hearing amplification. Compilation of their audiograms shows a consistent down-sloping sensorineural loss (Fig. 4B) with modest but consistent worsening across all frequencies.
Newborn hearing screening
Although the majority of patients were not referred to our practice immediately after birth, NHS information was available for 65% of individuals (34 of 52). Surprisingly, 50% of these (17 of 34) reported at least a unilateral pass on NHS; 11 of these reported bilateral passed screens. NT/NT individuals were more likely to have passed their NHS, which is not surprising given their eventual mild loss; however, several NT/T and T/T subjects had reportedly normal NHS results and subsequent severe-to-profound loss, suggesting a significant worsening of hearing (Fig. 4C). When individuals with a pass on NHS (suggesting hearing threshold <35 dB) and resulting hearing loss >40 dB PTA0.5–4 are included together with those with documented progression on serial audiometry, 28% (13 of 46) of all subjects with either serial audiograms or NHS results had findings suggestive of progressive hearing loss (Fig. 4D). This is consistent with our finding of 24% progression from serial audiometry alone.
DISCUSSION
Our examination of children with GJB2-associated hearing loss demonstrates the wide variety of phenotypes expressed in these individuals. Despite the relatively small number of children examined, the ethnic diversity of our patient population and availability of both audiometric and otolaryngologic records permitted the analysis of a variety of different genotypes with thorough phenotypic characterization. Consistent with previous large studies,10,11 individuals with genotypes consisting of two truncating mutations (T/T) had significantly more severe hearing loss than those with two nontruncating mutations (NT/NT), with NT/T genotypes expressing an intermediate phenotype. This is not surprising, given that a T/T genotype would be expected to yield no protein at all, whereas NT/NT individuals may express some poorly functional but possibly structurally intact gene product.
Nontruncating mutations present a diagnostic dilemma—how does one distinguish between a pathogenic nontruncating mutation and a benign polymorphism? Both have been described in the context of GJB2; although some variants, such as p.L90P, are universally acknowledged to be pathogenic, others, including p.V27I, are considered physiologic variants.6 When a sequence variant is first discovered, however, often in compound heterozygous combination with a known disease-causing allele such as c.35delG, it is not entirely certain whether the variant is truly the cause of hearing loss in that individual. More concrete attributions are based on epidemiologic studies comparing allele frequency in normal and affected populations,12 as well as on family studies, evolutionary conservation across different species, and in vitro functional studies of recombinant mutant proteins.20 However, such thorough evaluations have only been conducted on a handful of the mutations reported in the literature thus far. An in vitro study of connexin 26 conductance20 showed that many connexin 26 proteins with missense mutations are completely nonfunctional. However, one missense mutation, p.V84L, known to cause hearing loss in vivo, had normal function in vitro.20 Thus, even these in vitro studies may yield results that seem to conflict with in vivo clinical phenotypes.
Two common sequence changes in the Asian population, p.V27I and p.V37I, respectively yield functional and nonfunctional protein in vitro, in agreement with their clinical behavior.6 Both of these were detected frequently among our Asian patients, although individuals with p.V27I, a benign polymorphism, were not included in the study. p.V37I, despite its complete lack of function in vitro, was associated with only mild hearing loss when present in homozygosity or compound heterozygous combination with a truncating allele in this study. Overall, p.V37I and other nontruncating alleles accounted for 90% of the alleles identified among Asian children in our study; as a result, Asians overall had a relatively benign phenotype. In contrast, c.35delG and other truncating alleles accounted for 75% of the alleles identified among white and Latino children, who as a result were more hearing impaired as a group.
In particular, the p.V37I mutation illustrates the ethnic dichotomy in the literature and the diagnostic dilemma associated with mild phenotypes. Initially discovered in a Western study as a benign polymorphism,21 it was subsequently described instead as a pathogenic mutation.22 Studies from Asia have shown that it is carried in a significant fraction of certain East Asian populations, with allele frequencies up to 8.5% among Thai individuals23 and 11.6% in Taiwanese,24 suggesting once again that it may be benign. More in-depth analysis, however, including the in vitro functional experiments described above, has demonstrated that the p.V37I mutant protein is nonfunctional.25 Likewise, epidemiologic evaluation showed that the allele is indeed overrepresented among deaf individuals, indicating that it is most likely a pathogenic allele with variable penetrance.12,26 In particular, one study found that in patients with the p.V37I mutation, severity of hearing loss was correlated with the time elapsed since initial diagnosis; this was interpreted as indirect evidence for progressive hearing loss.12
From our direct serial audiometric data, we found that progressive hearing loss was common, with at least 24% of all studied patients showing evidence of progression. The relatively mild 9.0-dB threshold elevation likely underestimated the degree of worsening, given the artifactual tendency for audiometric indices to improve with a child's age owing simply to improved test sensitivity with better cooperation. Furthermore, the fraction of progressors was consistent at 28% on inclusion of children who passed their NHS and subsequently developed hearing loss. These findings are in rough agreement with progression rates of 17% and 22% reported previously.11,27
Progression was especially common among those in whom the p.V37I protein was the only connexin 26 protein produced, a population consisting of 11 children homozygous for p.V37I and 7 who were compound heterozygous for p.V37I and a second truncating allele. Thirty-nine percent of these children showed an average of 8.7 dB progression, with the majority requiring hearing amplification. Our evaluation was limited somewhat by the interval between audiograms, which was variable and insufficiently long to see large changes. Further follow-up of these individuals is thus warranted so that intervention is undertaken appropriately.
The high carrier frequency of p.V37I in Asian populations, its mild phenotype, which is often missed on NHS, and the tendency for progression mandate a high index of suspicion and close monitoring for any Asian children presenting with hearing loss. These features of p.V37I-associated hearing loss even suggest that there may be a degree of unrecognized hearing impairment among those Asian populations with high carrier frequency. Finally, despite the apparent complete lack of function of the p.V37I mutant protein in vivo, these results suggest that the protein may be in fact marginally functional but fails with time. As a gap junction protein involved in cellular homeostasis, such long-term mild dysfunction could manifest as progressive hearing loss and even as a predisposition to presbycusis.
Thus, our results and others suggest that there is considerable phenotypic variability for GJB2-associated hearing loss. The tendency toward slow but detectable progression in a significant fraction of children, especially those associated with the p.V37I allele, indicates the need for close audiometric follow-up in these individuals, with early intervention should it be clinically warranted. Further investigation will undoubtedly uncover more trends that will be helpful in the management of this complex clinical entity.
References
Mason JA, Herrmann KR . Universal infant hearing screening by automated auditory brainstem response measurement. Pediatrics 1998; 101: 221–228.
Parving A . The need for universal neonatal hearing screening—some aspects of epidemiology and identification. Acta Paediatr Suppl 1999; 88: 69–72.
Kelsell DP, Dunlop J, Stevens HP, et al. Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature 1997; 387: 80–83.
Kenneson A, Van Naarden Braun K, Boyle C . GJB2 (connexin 26) variants and nonsyndromic sensorineural hearing loss: a HuGE review. Genet Med 2002; 4: 258–274.
Nickel R, Forge A . Gap junctions and connexins in the inner ear: their roles in homeostasis and deafness. Curr Opin Otolaryngol Head Neck Surg 2008; 16: 452–457.
Ballana E, Ventayol M, Rabionet R, Gasparini P, Estivill X The connexin-deafness homepage, 2009. Available at: http://davinci.crg.es/deafness. Accessed April 16, 2009.
Richard G, Rouan F, Willoughby CE, et al. Missense mutations in GJB2 encoding connexin-26 cause the ectodermal dysplasia keratitis-ichthyosis-deafness syndrome. Am J Hum Genet 2002; 70: 1341–1348.
Zelante L, Gasparini P, Estivill X, et al. Connexin 26 mutations associated with the most common form of non-syndromic neurosensory autosomal recessive deafness (DFNB1) in Mediterraneans. Hum Mol Genet 1997; 6: 1605–1609.
Green GE, Scott DA, McDonald JM, Woodworth GG, Sheffield VC, Smith RJ . Carrier rates in the midwestern United States for GJB2 mutations causing inherited deafness. JAMA 1999; 281: 2211–2216.
Snoeckx RL, Huygen PL, Feldmann D, et al. GJB2 mutations and degree of hearing loss: a multicenter study. Am J Hum Genet 2005; 77: 945–957.
Marlin S, Feldmann D, Blons H, et al. GJB2 and GJB6 mutations: genotypic and phenotypic correlations in a large cohort of hearing-impaired patients. Arch Otolaryngol Head Neck Surg 2005; 131: 481–487.
Pollak A, Skórka A, Mueller-Malesinska M, et al. M34T and V37I mutations in GJB2 associated hearing impairment: evidence for pathogenicity and reduced penetrance. Am J Med Genet A 2007; 143: 2534–2543.
Denoyelle F, Marlin S, Weil D, et al. Clinical features of the prevalent form of childhood deafness, DFNB1, due to a connexin-26 gene defect: implications for genetic counseling. Lancet 1999; 353: 1298–1303.
Cohn ES, Kelley PM, Fowler TW, et al. Clinical studies of families with hearing loss attributable to mutations in the connexin 26 gene (GJB2/DFNB1). Pediatrics 1999; 103: 546–550.
Del Castillo I, Moreno-Pelayo MA, Del Castillo FJ, et al. Prevalence and evolutionary origins of the del(GJB6–D13S1830) mutation in the DFNB1 locus in hearing-impaired subjects: a multicenter study. Am J Hum Genet 2003; 73: 1452–1458.
Feldmann D, Denoyelle F, Chauvin P, et al. Large deletion of the GJB6 gene in deaf patients heterozygous for the GJB2 gene mutation: genotypic and phenotypic analysis. Am J Hum Genet A 2004; 127: 263–267.
Putcha GV, Bejjani BA, Bleoo S, et al. A multicenter study of the frequency and distribution of GJB2 and GJB6 mutations in a large North American cohort. Genet Med 2007; 9: 413–426.
Yotsumoto S, Hashiguchi T, Chen X, et al. Novel mutations in GJB2 encoding connexin-26 in Japanese patients with keratitis-ichthyosis-deafness syndrome. Br J Dermatol 2003; 4: 649–653.
Lerer I, Sagi M, Malamud E, Levi H, Raas-Rothschild A, Abeliovich D . Contribution of the connexin 26 mutations to nonsyndromic deafness in Ashkenazi patients and the variable phenotypic effect of the mutation 167delT. Am J Med Genet 2000; 95: 53–56.
Bruzzone R, Veronesi V, Gomès D, et al. Loss-of-function and residual channel activity of connexin 26 mutations associated with non-syndromic deafness. FEBS Lett 2003; 533: 79–88.
Kelley PM, Harris DJ, Comer BC, et al. Novel mutations in the connexin 26 gene (GJB2) that cause autosomal recessive (DFNB1) hearing loss. Am J Hum Genet 1998; 62: 792–799.
Rabionet R, Zelante L, López-Bigas N, et al. Molecular basis of childhood deafness resulting from mutations in the GJB2 (connexin 26) gene. Hum Genet 2000; 106: 40–44.
Wattanasirichaigoon D, Limwongse C, Jariengprasert C, et al. High prevalence of V37I genetic variant in the connexin-26 (GJB2) gene among non-syndromic hearing-impaired and control Thai individuals. Clin Genet 2004; 66: 452–460.
Hwa HL, Ko TM, Hsu CJ, et al. Mutation spectrum of the connexin 26 (GJB2) gene in Taiwanese patients with prelingual deafness. Genet Med 2003; 5: 161–165.
Palmada M, Schmalisch K, Böhmer C, et al. Loss of function mutations of the GJB2 gene detected in patients with DFNB1-associated hearing impairment. Neurobiol Dis 2006; 22: 112–118.
Huculak C, Bruyere H, Nelson TN, Kozak FK, Langlois S . V37I connexin 26 allele in patients with sensorineural hearing loss: evidence of its pathogenicity. Am J Med Genet A 2006; 140: 2394–2400.
Lee KH, Larson DA, Shott G, et al. Audiologic and temporal bone findings in patients with sensorineural hearing loss and GJB2 mutations. Laryngoscope 2009; 119: 554–558.
Acknowledgements
This work was performed with the support of the Department of Otolaryngology-Head and Neck Surgery and the Department of Pathology at Stanford University Hospital and Clinics.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Disclosure: The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Chan, D., Schrijver, I. & Chang, K. Connexin-26–associated deafness: Phenotypic variability and progression of hearing loss. Genet Med 12, 174–181 (2010). https://doi.org/10.1097/GIM.0b013e3181d0d42b
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1097/GIM.0b013e3181d0d42b
Keywords
This article is cited by
-
Therapeutic strategies targeting connexins
Nature Reviews Drug Discovery (2018)
-
A pipeline combining multiple strategies for prioritizing heterozygous variants for the identification of candidate genes in exome datasets
Human Genomics (2017)
-
The pathological effects of connexin 26 variants related to hearing loss by in silico and in vitro analysis
Human Genetics (2016)
-
Genetic Testing for Deaf and Hard of Hearing Individuals: Genetic Counseling
Current Genetic Medicine Reports (2016)
-
Inhibition of Connexin 26 by the AMP-Activated Protein Kinase
The Journal of Membrane Biology (2011)