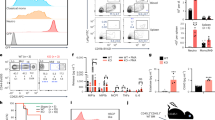
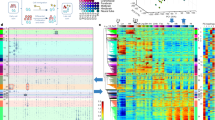
Abstract
We constructed and analyzed six serial analysis of gene expression (SAGE) libraries to identify genes with previously uncharacterized roles in spleen or thymus development. A total of 625 070 tags were sequenced from the three spleen (embryonic day (E)15.5, E16.5 and adult) and three thymus (E15.5, E18.5 and adult) libraries. These tags corresponded to 83 182 tag types, which mapped unambiguously to 36 133 different genes. Genes over-represented in these libraries, compared to 115 mouse SAGE libraries (www.mouseatlas.org), included genes of known and unknown immunological or developmental relevance. The expression profiles of 11 genes with unknown roles in spleen and thymus development were validated using reverse transcription-qPCR. We further characterized the expression of one of these candidates, RIKEN cDNA 9230105E10 that encodes a murine homolog of Trim5α, in numerous adult tissues and immune cell types. In addition, we demonstrate that transcript levels are upregulated in response to TLR stimulation of plasmacytoid dendritic cells and macrophages. This work provides the first evidence of regulated and cell type-specific expression of this gene. In addition, these observations suggest that the SAGE libraries provide an important resource for further investigations into the molecular mechanisms regulating spleen and thymus organogenesis, as well as the development of immunological competence.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 digital issues and online access to articles
$119.00 per year
only $19.83 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lu J, Chang P, Richardson JA, Gan L, Weiler H, Olson EN . The basic helix–loop–helix transcription factor capsulin controls spleen organogenesis. Proc Natl Acad Sci USA 2000; 97: 9525–9530.
Hecksher-Sorensen J, Watson RP, Lettice LA, Serup P, Eley L, De Angelis C et al. The splanchnic mesodermal plate directs spleen and pancreatic laterality, and is regulated by Bapx1/Nkx3.2. Development 2004; 131: 4665–4675.
Dear TN, Colledge WH, Carlton MB, Lavenir I, Larson T, Smith AJ et al. The Hox11 gene is essential for cell survival during spleen development. Development 1995; 121: 2909–2915.
Tribioli C, Lufkin T . The murine Bapx1 homeobox gene plays a critical role in embryonic development of the axial skeleton and spleen. Development 1999; 126: 5699–5711.
Kanzler B, Dear TN . Hox11 acts cell autonomously in spleen development and its absence results in altered cell fate of mesenchymal spleen precursors. Dev Biol 2001; 234: 231–243.
Cyster JG . Lymphoid organ development and cell migration. Immunol Rev 2003; 195: 5–14.
Mebius RE . Organogenesis of lymphoid tissues. Nat Rev Immunol 2003; 3: 292–303.
Boehm T, Bleul CC, Schorpp M . Genetic dissection of thymus development in mouse and zebrafish. Immunol Rev 2003; 195: 15–27.
Gill J, Malin M, Sutherland J, Gray D, Hollander G, Boyd R . Thymic generation and regeneration. Immunol Rev 2003; 195: 28–50.
Bockman DE . Development of the thymus. Microsc Res Technol 1997; 38: 209–215.
Coffer PJ, Burgering BM . Forkhead-box transcription factors and their role in the immune system. Nat Rev Immunol 2004; 4: 889–899.
Gordon J, Bennett AR, Blackburn CC, Manley NR . Gcm2 and Foxn1 mark early parathyroid- and thymus-specific domains in the developing third pharyngeal pouch. Mech Dev 2001; 103: 141–143.
Su D, Ellis S, Napier A, Lee K, Manley NR . Hoxa3 and pax1 regulate epithelial cell death and proliferation during thymus and parathyroid organogenesis. Dev Biol 2001; 236: 316–329.
Manley NR, Capecchi MR . The role of Hoxa-3 in mouse thymus and thyroid development. Development 1995; 121: 1989–2003.
Su DM, Manley NR . Hoxa3 and pax1 transcription factors regulate the ability of fetal thymic epithelial cells to promote thymocyte development. J Immunol 2000; 164: 5753–5760.
Wallin J, Eibel H, Neubuser A, Wilting J, Koseki H, Balling R . Pax1 is expressed during development of the thymus epithelium and is required for normal T-cell maturation. Development 1996; 122: 23–30.
Hetzer-Egger C, Schorpp M, Haas-Assenbaum A, Balling R, Peters H, Boehm T . Thymopoiesis requires Pax9 function in thymic epithelial cells. Eur J Immunol 2002; 32: 1175–1181.
Dallman MJ, Smith E, Benson RA, Lamb JR . Notch: control of lymphocyte differentiation in the periphery. Curr Opin Immunol 2005; 17: 259–266.
Robey EA, Bluestone JA . Notch signaling in lymphocyte development and function. Curr Opin Immunol 2004; 16: 360–366.
Radtke F, Wilson A, Mancini SJ, MacDonald HR . Notch regulation of lymphocyte development and function. Nat Immunol 2004; 5: 247–253.
Guidos CJ . Notch signaling in lymphocyte development. Semin Immunol 2002; 14: 395–404.
Allman D, Aster JC, Pear WS . Notch signaling in hematopoiesis and early lymphocyte development. Immunol Rev 2002; 187: 75–86.
Anderson AC, Robey EA, Huang YH . Notch signaling in lymphocyte development. Curr Opin Genet Dev 2001; 11: 554–560.
Wahl MB, Heinzmann U, Imai K . LongSAGE analysis significantly improves genome annotation: identifications of novel genes and alternative transcripts in the mouse. Bioinformatics 2005; 21: 1393–1400.
Saha S, Sparks AB, Rago C, Akmaev V, Wang CJ, Vogelstein B et al. Using the transcriptome to annotate the genome. Nat Biotechnol 2002; 20: 508–512.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25: 25–29.
Wang VE, Tantin D, Chen J, Sharp PA . B cell development and immunoglobulin transcription in Oct-1-deficient mice. Proc Natl Acad Sci USA 2004; 101: 2005–2010.
Messier H, Brickner H, Gaikwad J, Fotedar A . A novel POU domain protein which binds to the T-cell receptor beta enhancer. Mol Cell Biol 1993; 13: 5450–5460.
Sakaguchi S . Naturally arising Foxp3-expressing CD25+CD4+ regulatory T cells in immunological tolerance to self and non-self. Nat Immunol 2005; 6: 345–352.
Fontenot JD, Rudensky AY . A well adapted regulatory contrivance: regulatory T cell development and the forkhead family transcription factor Foxp3. Nat Immunol 2005; 6: 331–337.
Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY . Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity 2005; 22: 329–341.
Ochs HD, Ziegler SF, Torgerson TR . FOXP3 acts as a rheostat of the immune response. Immunol Rev 2005; 203: 156–164.
Audic S, Claverie JM . The significance of digital gene expression profiles. Genome Res 1997; 7: 986–995.
Smith E, Sigvardsson M . The roles of transcription factors in B lymphocyte commitment, development, and transformation. J Leukocyte Biol 2004; 75: 973–981.
Barrans SL, Fenton JA, Banham A, Owen RG, Jack AS . Strong expression of FOXP1 identifies a distinct subset of diffuse large B-cell lymphoma (DLBCL) patients with poor outcome. Blood 2004; 104: 2933–2935.
Wlodarska I, Veyt E, De Paepe P, Vandenberghe P, Nooijen P, Theate I et al. FOXP1, a gene highly expressed in a subset of diffuse large B-cell lymphoma, is recurrently targeted by genomic aberrations. Leukemia 2005; 19: 1299–1305.
Tarlinton D, Light A, Metcalf D, Harvey RP, Robb L . Architectural defects in the spleens of Nkx2-3-deficient mice are intrinsic and associated with defects in both B cell maturation and T cell-dependent immune responses. J Immunol 2003; 170: 4002–4010.
Hahm K, Sum EY, Fujiwara Y, Lindeman GJ, Visvader JE, Orkin SH . Defective neural tube closure and anteroposterior patterning in mice lacking the LIM protein LMO4 or its interacting partner Deaf-1. Mol Cell Biol 2004; 24: 2074–2082.
Wang Y, Wimmer U, Lichtlen P, Inderbitzin D, Stieger B, Meier PJ et al. Metal-responsive transcription factor-1 (MTF-1) is essential for embryonic liver development and heavy metal detoxification in the adult liver. FASEB J 2004; 18: 1071–1079.
Bardoux P, Zhang P, Flamez D, Perilhou A, Lavin TA, Tanti JF et al. Essential role of chicken ovalbumin upstream promoter-transcription factor II in insulin secretion and insulin sensitivity revealed by conditional gene knockout. Diabetes 2005; 54: 1357–1363.
You LR, Lin FJ, Lee CT, DeMayo FJ, Tsai MJ, Tsai SY . Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature 2005; 435: 98–104.
Allen III RD, Bender TP, Siu G . c-Myb is essential for early T cell development. Genes Dev 1999; 13: 1073–1078.
Bender TP, Kremer CS, Kraus M, Buch T, Rajewsky K . Critical functions for c-Myb at three checkpoints during thymocyte development. Nat Immunol 2004; 5: 721–729.
Lieu YK, Kumar A, Pajerowski AG, Rogers TJ, Reddy EP . Requirement of c-myb in T cell development and in mature T cell function. Proc Natl Acad Sci USA 2004; 101: 14853–14858.
Cai S, Han HJ, Kohwi-Shigematsu T . Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat Genet 2003; 34: 42–51.
Nie H, Maika SD, Tucker PW, Gottlieb PD . A role for SATB1, a nuclear matrix association region-binding protein, in the development of CD8SP thymocytes and peripheral T lymphocytes. J Immunol 2005; 174: 4745–4752.
Leiden JM . Transcriptional regulation of T cell receptor genes. Annu Rev Immunol 1993; 11: 539–570.
Rothenberg EV, Taghon T . Molecular genetics of T cell development. Annu Rev Immunol 2005; 23: 601–649.
Su DM, Navarre S, Oh WJ, Condie BG, Manley NR . A domain of Foxn1 required for crosstalk-dependent thymic epithelial cell differentiation. Nat Immunol 2003; 4: 1128–1135.
Meng A, Moore B, Tang H, Yuan B, Lin S . A Drosophila doublesex-related gene, terra, is involved in somitogenesis in vertebrates. Development 1999; 126: 1259–1268.
Kim S, Kettlewell JR, Anderson RC, Bardwell VJ, Zarkower D . Sexually dimorphic expression of multiple doublesex-related genes in the embryonic mouse gonad. Gene Expr Patterns 2003; 3: 77–82.
Azakie A, Lamont L, Fineman JR, He Y . Divergent transcriptional Enhancer Factor 1 (DTEF-1) regulates the cardiac Troponin T promoter. Am J Physiol Cell Physiol 2005; 289: C1522–C1534.
Brendolan A, Ferretti E, Salsi V, Moses K, Quaggin S, Blasi F et al. A Pbx1-dependent genetic and transcriptional network regulates spleen ontogeny. Development 2005; 132: 3113–3126.
Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y et al. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-beta gene regulatory elements. Cell 1988; 54: 903–913.
Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, Watanabe N et al. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell 1993; 75: 83–97.
Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J . The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 2004; 427: 848–853.
Yap MW, Nisole S, Lynch C, Stoye JP . Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc Natl Acad Sci USA 2004; 101: 10786–10791.
Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L et al. The tripartite motif family identifies cell compartments. EMBO J 2001; 20: 2140–2151.
Iwasaki A, Medzhitov R . Toll-like receptor control of the adaptive immune responses. Nat Immunol 2004; 5: 987–995.
Verthelyi D, Zeuner RA . Differential signaling by CpG DNA in DCs and B cells: not just TLR9. Trends Immunol 2003; 24: 519–522.
Vogel SN, Fitzgerald KA, Fenton MJ . TLRs: differential adapter utilization by toll-like receptors mediates TLR-specific patterns of gene expression. Mol Interven 2003; 3: 466–477.
Beutler B . Inferences, questions and possibilities in Toll-like receptor signalling. Nature 2004; 430: 257–263.
Beutler B, Du X, Poltorak A . Identification of Toll-like receptor 4 (Tlr4) as the sole conduit for LPS signal transduction: genetic and evolutionary studies. J Endotoxin Res 2001; 7: 277–280.
Dunne A, O’Neill LA . Adaptor usage and Toll-like receptor signaling specificity. FEBS Lett 2005; 579: 3330–3335.
Hirschfeld M, Ma Y, Weis JH, Vogel SN, Weis JJ . Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine toll-like receptor 2. J Immunol 2000; 165: 618–622.
Dinel S, Bolduc C, Belleau P, Boivin A, Yoshioka M, Calvo E et al. Reproducibility, bioinformatic analysis and power of the SAGE method to evaluate changes in transcriptome. Nucleic Acids Res 2005; 33: e26.
Ewing B, Green P . Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 1998; 8: 186–194.
Pfaffl MW . A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29: e45.
Taylor PA, Panoskaltsis-Mortari A, Swedin JM, Lucas PJ, Gress RE, Levine BL et al. L-Selectin(hi) but not the L-selectin(lo) CD4+25+ T-regulatory cells are potent inhibitors of GVHD and BM graft rejection. Blood 2004; 104: 3804–3812.
Acknowledgements
Eosinophil and mast cell RNA was a kind gift from BH Koller, CD25− and Treg cell RNA were a kind gift from J Serody and purified B cells were a kind gift of SH Clarke. This work was carried out as part of the Mouse Atlas of Gene Expression Project, a collaborative project involving the following Principle Investigators: Cheryl D Helgason, Pamela Hoodless, Steven Jones, Marco Marra, Elizabeth M Simpson and Greg Riggins. Daniella Gerhardt and Robert Strausberg were also important contributors to this project. We would like to acknowledge members of the Genome Sciences Center sequencing, bioinformatics and SAGE library construction teams, in particular Anita Charters, Jennifer Asano, Allen Delaney, Jaswinder Khattra and Asim Siddiqui, for their valuable contributions. We would also like to thank Adrian Burke and Caroline Astell for their assistance in project management and the staff of the Joint Animal Facility at the BBCRC for maintaining the mice. Funding for this project was provided by Genome Canada through Genome BC (CDH) and the National Institute of Health AI29564 (JT), with infrastructure support provided by the BC Cancer Foundation. Kristi L Williams is supported in part by the 2004 Amgen/Focis Fellowship Award, whereas Cheryl D Helgason is a scholar of the Canadian Institutes of Health Research and the Michael Smith Foundation for Health Research.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary information accompanies the paper on Gene and Immunity website (http://www.nature.com/gene)
Supplementary information
Rights and permissions
About this article
Cite this article
Hoffman, B., Williams, K., Tien, A. et al. Identification of novel genes and transcription factors involved in spleen, thymus and immunological development and function. Genes Immun 7, 101–112 (2006). https://doi.org/10.1038/sj.gene.6364270
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.gene.6364270