Abstract
The island population of Tristan da Cunha has a well-documented genealogy that dates to its first permanent settlement in 1816. The current population is thought to have descended from only seven females and eight males. Today, there are seven family names in use, corresponding to the number of founding fathers with present-day male descendents. Y chromosome polymorphisms have previously been shown to be reliable tools for tracing patrilineal genealogies. Here, we studied Y chromosome polymorphisms in a sample from Tristan da Cunha together with genealogical records to (i) infer the haplotypes of the seven founders and (ii) test if the Y chromosome transmission is consistent with the documented patrilineal history of the island community. We observed nine Y chromosome haplotypes of which seven could be traced to the known ancestors. Of the two additional lineages, one probably evolved from a founder haplotype due to a single-step microsatellite mutation, while the other had an obvious non-island origin. Its introduction, however, is not reflected in the records. Four more instances of non-paternity were identified, with the ‘new’ chromosomes matching other island haplotypes. The Y chromosome data presented here question the validity of some of the genealogical documentation and emphasise the value of genetic studies in tracing ancestry.
Similar content being viewed by others
Introduction
Genealogy is the discipline that traditionally deals with the reconstruction of family history. So far, the quest to trace ancestors and distant relatives has mainly relied on oral traditions, church records and other documents. The human genome constitutes an archive of our history and evolutionary past that can be explored for genealogical research. Genetic markers, like mitochondrial DNA (mtDNA) and Y chromosome polymorphisms, have been shown in historical investigations to be reliable tools for tracing matri- and patrilineal genealogies, respectively.1,2 Since hereditary family names are passed down in the same way as paternally inherited Y chromosomes, a surname should, within a genealogy, correlate with a Y chromosome haplotype.3,4
The island population of Tristan da Cunha is unique with regard to its well-documented genealogy that dates back to the first permanent settlement in the early 19th century.5 The current population of 278 individuals6 is thought to have descended from 15 ancestors, seven females and eight males who arrived on the island at various times between 1816 and 1908. The male founders were all of western European ancestry, originating from Scotland, England, Holland, the USA and Italy. Today, there are seven family names in use (Glass, Green, Hagan, Lavarello, Repetto, Rogers and Swain), corresponding to the number of founding fathers with present-day male descendents. In addition to these known donors, there is also evidence for ‘hidden ancestors’ who supposedly contributed to the gene pool, but took their names with them upon leaving the island.7
In a previous study, the present-day mtDNA pool of the Tristan islanders was traced to five female ancestors.8 These data identified inconsistencies in the number of mtDNA types and the written records. Although the historical documents mentioned two sister pairs among the founding females, mtDNA data showed support for only one pair of sisters. In the present study, we studied 13 Y chromosome polymorphisms in a sample from Tristan da Cunha together with the available genealogical information to (i) infer the Y chromosome haplotypes of the known male founders who brought their surnames to the island and (ii) test if the Y chromosome transmission is consistent with the documented patrilineal history of the island community.
Subjects and methods
DNA samples were prepared from peripheral blood by standard phenol–chloroform procedures from 76 males in a sample collected on the island of Tristan da Cunha in 1982. This study was approved by the Committee for Research on Human Subjects at the University of Witwatersrand, Johannesburg, South Africa. Out of respect for the sensitivities of the present inhabitants of Tristan da Cunha, the observed haplotypes were neither associated with the family names nor with the origins of the founding fathers in order to avoid identification. DNA samples were screened for 13 Y chromosome-specific polymorphisms. The seven bi-allelic Y chromosome markers used here (Figure 1) have previously been shown to define haplogroups frequently found in European populations.9,10,11 Haplogroup designation follows the nomenclature proposed by the Y Chromosome Consortium (YCC).12 The markers M1 (Y chromosome Alu insertion polymorphism ‘YAP’) and M9 were typed in all samples. The allelic states of the remaining polymorphisms were determined hierarchically based on the known Y chromosome genealogy (Figure 1).12 M1, p12f2 and SRY10831 were screened as described.13,9,14 M9,15 M170, M207, M21316 were typed using agarose gel-based PCR–RFLP assays. The protocols for PCR amplification and the detection of polymorphic markers are available from the authors upon request. The six microsatellite loci DYS19, DYS388, DYS390, DYS391, DYS392 and DYS39317 were amplified in two separate multiplex PCR reactions. The PCR conditions are available from the authors upon request. The fluorescently labelled PCR products were resolved on polyacrylamide gels in an automated ABI 377 DNA sequencer. The number of repeats at each locus was determined adopting the proposed nomenclature.18 Haplotypes were defined by both bi-allelic and microsatellite polymorphisms. Microsatellite haplotypes were constructed using the number of repeats (alleles) in the order DYS19-DYS388-DYS390-DYS391-DYS392-DYS393.
Y chromosome phylogeny of the bi-allelic poly-morphisms used in the study (black bars). The three Y chromosome haplogroups found in the sample from Tristan da Cunha are indicated (solid lines). Haplogroups are designated according to the YCC nomenclature.12
Results and discussion
We derived nine Y chromosome haplotypes of which seven could unambiguously be traced to the seven men who are known to have introduced surnames to the island (Table 1, haplotypes shown in bold). These seven haplotypes segregated either on a haplogroup R-M207 (× SRY10831.2) or an I-M170 background. R-M207 (x SRY10831.2) is mainly equivalent to R-M173 (× M17),12 which is the dominant Y chromosome lineage in western European populations where it reaches frequencies of 50% and above.10,11 Haplogroup I-M170 is virtually confined to Europe. In western Europeans it has been observed at moderate frequencies (eg 22% in Dutch and 8% in Italians).10,11 Hence, the presence of haplogroups R-M207 and I-M170 in the sample from Tristan da Cunha is consistent with the western European ancestry of the known founding fathers, as suggested by historical records.7
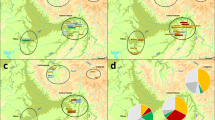
Moreover, two additional haplotypes were discovered (Table 1, haplotypes in families 3 and 4 shown in italics). The first one was found in family 3, in four individuals spanning three generations (Figure 2a). This haplotype could have evolved from one of the two haplotypes already present in families 3 and 5 (Table 1), because of single-step microsatellite mutations at either DYS391 or DYS392. Since the observed mutation rate at DYS392 is very low in European populations (0 in 415 meioses, 95% CI 7.15 × 10−3),19 the mutation at DYS391 seems to be the most likely event that gave rise to the new haplotype through transmission within family 3. Unfortunately, we do not have DNA from the parental generation where the mutation is presumed to have occurred, making paternity testing using autosomal markers impossible. Assuming the origin of the new haplotype within family 3, we estimated the mutation rate at the DYS391 locus to be 1 in 100 meioses (10 × 10−3, 95% CI 0.2–54.5 × 10−3, excluding inconsistent transmissions, discussed later in the text). The mutation rate at this locus determined in a random sample of father/son pairs was reported to be 4.82 × 10−3 (95% CI 0.61–16.4 × 10−3).19
Partial male genealogies showing inconsistencies in the transmission of Y chromosome haplotypes in five of the seven families on Tristan da Cunha. To conceal the identities of the families, only partial sib-ships are shown without indicating the generations in the pedigrees. Not all the males from whom Y chromosome data were obtained are presented. Tested individuals are indicated by filled squares. (a) Partial genealogy of family 3. An additional haplotype was found in four individuals spanning three generations. This haplotype most likely evolved from the haplotype associated with family 3 due to a single-step microsatellite mutation at DYS391. Arrow indicates the individual in whom the mutation probably originated. (b) Partial genealogy of family 4. The additional haplotype has a non-island origin (see Table 1). (c) Partial genealogy of family 5. The additional haplotypes observed in two branches of the genealogy are identical to the haplotype of family 7. (d) Partial genealogy of family 6. The additional haplotype in one branch of the genealogy is identical to the haplotype associated with family 5. (e) Partial genealogy of family 7. The additional haplotype is identical to the haplotype associated with family 5.
The second additional haplotype was observed in family 4, in one individual who had no sons (Figure 2b). This haplotype can be distinguished from all the others in the Tristan sample due to a reversion at SRY10831 on a haplogroup R-M207 background (Figure 1 and Table 1). This mutation defines a lineage that is particularly frequent in eastern and northern Europeans (54% in Polish, 47% in Russians, 31% in Norwegians).9 Furthermore, the microsatellite allele constellation found in the Tristan individual (Table 1) represents the most common haplotype within this haplogroup.20 Based on genealogical information, we deduced that this new haplotype was introduced into family 4 in the early 1900s. It is well documented that many passenger ships, cargo vessels and whalers, some from Russia and Norway, used the island as a stopover port for trade and replenishing supplies.7 Perhaps the new lineage was brought to the island during such a visit. This finding provides evidence for the contribution of a hidden ancestor who left his genes but not his name on the island.
The Y chromosome data in conjunction with genealogical information enabled us to unequivocally identify four additional instances of nonpaternity (in families 5–7; Table 1, haplotypes shown in regular font). In these cases, Y chromosomes were introduced that belonged to a different haplogroup from that associated with the family or surname. These ‘newcomers’, however, matched two other lineages already present on the island (Table 1 and Figures 2c–e). The ‘new’ chromosomes observed in two branches of the genealogy in family 5 are identical to the haplotype of family 7 (Figure 2c). The haplotype associated with family 5, on the other hand, was also found in families 6 (Figure 2d) and 7 (Figure 2e). There are two possible explanations for these findings: First, the introduced chromosomes could be the result of pre- or extramarital affairs involving men from the island community. There are indications in the records that would support this possibility.7 The second explanation, of course, could be that the chromosomes were left on the island by other hidden ancestors who happened to have haplotypes that were identical to those of some of the islanders. In fact, the ‘new’ microsatellite haplotypes appear to be common in European populations; in the European Y-STR Haplotype Reference Database21 the frequency for the haplotype of family 5 has been reported as 1.5 × 10−3 and for that of family 7 as 3.9 × 10−2 (the haplotype frequencies are calculated using five microsatellite loci only without considering DYS388 and the bi-allelic markers. As a result, the actual frequencies of the extended haplotypes used in this study would be lower). It is difficult to assess the probability of an internal versus an external contribution of these chromosomes to the Tristan population. However, given the presence of the two lineages in the community already (in families 5 and 7) and the geographic remoteness of Tristan da Cunha (thus the limited traffic to and from the island; Figure 3), it seems improbable that mysterious visitors contributed their chromosomes in all four instances.
We calculated the incidence of non-paternity in the male Tristan sample to be about 4% (five instances in 118 male births; illegitimate daughters cannot be detected studying Y chromosome transmission). Although this rate falls within the observed range of estimates (between 1.3 and 30%),3,22 it is most likely underestimated, given the high level of inbreeding7 and the consequently high extent of Y chromosome haplotype sharing on the island. Obviously, we could only discern those cases in which the Y chromosomes of the biological father and the name-giver were different. Interestingly, a study based on 27 autosomal serogenetic markers, including the highly informative Gm haplotype, did not indicate any inconsistencies in the genealogies of the Tristan da Cunha islanders.23
The uniqueness of the Tristan genealogy covering a period of almost 200 years presented us with an ideal opportunity of testing the accuracy of written records with information stored in DNA. The Y chromosome data shown here, together with those based on mtDNA,8 question the validity of some of the genealogical documentation. This study demonstrates convincingly that DNA is a powerful archival resource that can be used to test questions and to refine theories pertaining to human history and origins.
References
Gill P, Ivanov PL, Kimpton C et al: Identification of the remains of the Romanov family by DNA analysis. Nature Genet 1994; 6: 130–135.
Foster EA, Jobling MA, Taylor PG et al: Jefferson fathered slave's last child. Nature 1998; 396: 27–28.
Sykes B, Irven C : Surnames and the Y chromosome. Am J Hum Genet 2000; 66: 1417–1419.
Jobling MA : In the name of the father: surnames and genetics. Trends Genet 2001; 17: 353–357.
Roberts DF : Genetic effects of population size reduction. Nature 1968; 220: 1084–1088.
Department for Environment, Food and Rural Affairs (DEFRA), UK Overseas Territories, consensus data for 2001, http://www.defra.gov.uk/science/geneticresources/access/overseas.htm.
Roberts DF : The development of inbreeding in an island population; in Morris LN (ed): Human populations, genetic variation and evolution. San Francisco, Chandler Publishing Company, 1971, pp 381–391.
Soodyall H, Jenkins T, Mukherjee A, du Toit E, Roberts DF, Stoneking M : The founding mitochondrial DNA lineages of Tristan da Cunha islanders. Am J Phys Anthropol 1997; 104: 157–166.
Rosser ZH, Zerjal T, Hurles ME et al: Y-chromosomal diversity in Europe is clinal and influenced by geography, rather than by language. Am J Hum Genet 2000; 67: 1526–1543.
Semino O, Passarino G, Oefner PJ et al: The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 2000; 290: 1155–1159.
Underhill PA, Shen P, Jin L et al: Y chromosome sequence variation and the history of human populations. Nature Genet 2000; 26: 358–361.
YCC – the Y Chromosome Consortium. A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 2002; 12: 339–348.
Hammer MF, Horai S : Y chromosomal variation and the peopling of Japan. Am J Hum Genet 1995; 56: 951–962.
Santos FR, Pandaya A, Tyler-Smith C et al: The central Siberian origin for native American Y chromosomes. Am J Hum Genet 1999; 64: 619–628.
Underhill PA, Jin L, Lin AA et al: Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res 1997; 7: 996–1005.
Shen P, Wang F, Underhill P et al: Population genetic implications from sequence variation in four Y chromosome genes. Proc Natl Acad Sci USA 2000; 97: 7354–7359.
Jobling MA, Tyler-Smith C: Fathers and sons: the Y chromosome and human evolution. Trends Genet 1995; 11: 449–456.
Kayser M, Caglia A, Corach D et al: Evaluation of Y-chromosomal STRs: a multicenter study. Int J Legal Med 1997; 110: 125–133.
Kayser M, Roewer L, Hedman M et al: Characteristics and frequency of germline mutations at microsatellite loci from the human Y chromosome, as revealed by direct observation in father/son pairs. Am J Hum Genet 2000; 66: 1580–1588.
Zerjal T, Pandya A, Santos FR et al: The use of Y-chromosomal DNA variation to investigate population history: recent male spread in Asia and Europe. in Papiha SS, Deka R, Chakraborty R (eds): Genomic diversity: applications in human population genetics. New York, Plenum Press, 1999, pp 91–101.
Y-STR Haplotype Reference Database, http://ystr.charite.de
MacIntyre S, Sooman A : Non-paternity and prenatal genetic screening. Lancet 1991; 338: 869–871.
Jenkins T, Beighton P, Steinberg AG : Serogenetic studies on the inhabitants of Tristan da Cunha. Ann Hum Genet 1985; 12: 363–371.
Acknowledgements
We would like to thank Professor E du Toit for samples, Professor D Roberts for genealogical data as well as Ms C Dorf for technical assistance. This research was supported by grants from the National Health Laboratory Service (formerly known as South African Institute for Medical Research), the South African Medical Research Council, the National Research Foundation and the University of the Witwatersrand. AN is the recipient of the Hillel Friedland Postdoctoral Research Fellowship from the University of the Witwatersrand.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Soodyall, H., Nebel, A., Morar, B. et al. Genealogy and genes: tracing the founding fathers of Tristan da Cunha. Eur J Hum Genet 11, 705–709 (2003). https://doi.org/10.1038/sj.ejhg.5201022
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201022
Keywords
This article is cited by
-
Three hundred years of low non-paternity in a human population
Heredity (2015)
-
Temporal differentiation across a West-European Y-chromosomal cline: genealogy as a tool in human population genetics
European Journal of Human Genetics (2012)
-
Significant genetic differentiation between Poland and Germany follows present-day political borders, as revealed by Y-chromosome analysis
Human Genetics (2005)
-
Genetic ancestry and the search for personalized genetic histories
Nature Reviews Genetics (2004)