Abstract
Sarcomatoid mesothelioma, a histological subtype of malignant pleural mesothelioma, is a very aggressive tumor with a poor prognosis. Histological diagnosis of sarcomatoid mesothelioma largely depends on the histomorphological feature of spindled tumor cells with immunohistochemical reactivity to cytokeratins. Diagnosis also requires clinico-radiological and/or macroscopic evidence of an extrapulmonary location to differentiate it from lung sarcomatoid carcinoma. Although there are promising immunohistochemical antibody panels to differentiate mesothelioma from lung carcinoma, a consensus on the immunohistochemical markers that distinguish sarcomatoid mesothelioma from lung sarcomatoid carcinoma has not been reached and requires further study. We performed whole gene expression analysis of formalin-fixed paraffin-embedded tissue from sarcomatoid mesothelioma and lung sarcomatoid carcinoma and observed significant differences in the expression of MUC4 and other genes between sarcomatoid mesothelioma and lung sarcomatoid carcinoma. Immunohistochemistry demonstrated that MUC4 was expressed in the spindled tumor cells of lung sarcomatoid carcinoma (21/29, 72%) but was not expressed in any sarcomatoid mesothelioma (0/31, 0%). To differentiate sarcomatoid mesothelioma from lung sarcomatoid carcinoma, negative MUC4 expression showed 100% sensitivity and 72% specificity and accuracy rate of 87%, which is higher than immunohistochemical markers such as calretinin, D2-40 and Claudin-4. Therefore, we recommend to include MUC4 as a novel and useful negative immunohistochemical marker for differentiating sarcomatoid mesothelioma from lung sarcomatoid carcinoma.
Similar content being viewed by others
Main
Malignant pleural mesothelioma, a highly aggressive tumor with a poor prognosis, is strongly associated with asbestos exposure; its incidence is increasing in Japan and Western countries and is expected to increase in developing countries.1 It is histologically classified into three subtypes: epithelioid, biphasic, and sarcomatoid mesothelioma.2 The International Mesothelioma Interest Group (IMIG) has published guidelines for the differential diagnosis of epithelioid mesothelioma from lung adenocarcinoma and squamous cell carcinoma using immunohistochemical antibody panels of mesothelioma markers (calretinin, D2-40, WT1, cytokeratin 5/6), lung adenocarcinoma markers (CEA, TTF-1, Napsin-A, MOC-31, BerEP4, BG8, B72.3) and lung squamous carcinoma markers (p63, p40, MOC-31, Ber-EP4, cytokeratin 5/6).3
However, a consensus on the immunohistochemical markers that differentiate sarcomatoid mesothelioma from lung sarcomatoid carcinoma has not been reached and requires further study. The histological diagnosis of sarcomatoid mesothelioma largely depends on the histomorphological feature of spindled tumor cells supported by immunohistochemical cytokeratin reactivity; it also requires clinico-radiological and/or macroscopic evidence of an extrapulmonary location. The immunohistochemical markers for lung adenocarcinoma and squamous carcinoma are not useful for diagnosing lung sarcomatoid carcinoma. To date, D2-40 and calretinin are two commonly used positive mesothelial markers expressed in sarcomatoid mesothelioma.4, 5, 6, 7 However, without convincing calretinin and D2-40 positivity, it is difficult to differentiate sarcomatoid mesothelioma from sarcomatoid carcinoma. In previous reports, including ours, high D2-40 sensitivity has been reported to differentiate sarcomatoid mesothelioma from lung sarcomatoid carcinoma; however, D2-40 specificity is not perfect.6, 7 Therefore, the clinico-radiological identification of tumor location at the extrapulmonary site remains essential to differentiate between these two diseases.
In recent decades, gene expression profiling has been used in many cancers to identify the pathways involved in malignant transformation and to identify novel candidate diagnostic and prognostic markers. We have recently reported the application of gene expression analysis to identify novel markers differentiating epithelioid mesothelioma from reactive mesothelial hyperplasia by PCR array.8 Although gene expression analysis requires specimens with a high proportion of tumor cells containing good quality RNA, we successfully analyzed the RNA extracted from formalin-fixed paraffin-embedded samples.
The aim of this study was to perform gene expression analysis on spindled tumor cells dissected from formalin-fixed paraffin-embedded tissue of sarcomatoid mesothelioma and lung sarcomatoid carcinoma. Our gene expression microarray data identified several novel genes that are differentially expressed between sarcomatoid mesothelioma and lung sarcomatoid carcinoma, and of these, we validated MUC4 as a novel and useful negative immunohistochemical marker differentiating sarcomatoid mesothelioma from lung sarcomatoid carcinoma.
Materials and methods
Formalin-Fixed Paraffin-Embedded Tissue Samples
Sarcomatoid mesothelioma and lung sarcomatoid carcinoma cases were retrieved from surgical pathology archives of our department during 2005–2014. The clinical details were also reviewed from the patient record files. The location of tumor was confirmed by reviewing clinical information (especially chest computed tomography findings to confirm the tumor localization), gross findings and reviewing histological sections stained with H&E and Elastica van Gieson. All lung sarcomatoid carcinoma cases in this study were located in the pulmonary parenchyma, which was confirmed by radiological, thoracoscopic and operative findings. None of the lung sarcomatoid carcinoma showed diffuse pleurotropic growth pattern described as ‘pseudomesotheliomatous growth’. Sarcomatoid mesothelioma was located in extrapulmonary site showing dominant pleurotrophic growth pattern without obvious tumor mass in lung parenchyma. Pathological diagnosis of each case was confirmed by histological findings and immunohistochemical marker panel recommended by Guidelines for Pathologic Diagnosis of Malignant Mesothelioma-2012 Update of the Consensus Statement from the International Mesothelioma Interest Group3 and current 2015 WHO histological classification of tumours of the lung, pleura, thymus and heart.9 Sarcomatoid mesothelioma is characterized by a proliferation of spindle cells arranged in fascicles or having a haphazard distribution involving adjacent adipose tissue, parietal pleura or lung parenchyma.9 Lung sarcomatoid carcinoma is a poorly differentiated non-small cell lung carcinoma that contains a component of sarcoma or sarcoma-like (spindle and/or giant cell) differentiation. Lung sarcomatoid carcinoma is a group of five types of carcinomas based on specific histological criteria and described as giant cell carcinoma, pleomorphic carcinoma, carcinosarcoma, spindle cell carcinoma and pulmonary blastoma. Of these, spindle cell carcinoma and pleomorphic carcinoma with predominant spindle cell component requires the differentiation from sarcomatoid mesothelioma. The number of patients who were diagnosed as sarcomatoid mesothelioma and lung sarcomatoid carcinoma after surgical resection and/or autopsy examination in Hiroshima University Hospital during 2005–2014 were 35 and 34 respectively, suggesting similar frequencies of their incidence. Localization of four cases of sarcomatoid mesothelioma and five cases of lung sarcomoid carcinoma could not be confirmed and thus were excluded from this study. Finally, 31 cases of sarcomatoid mesothelioma and 29 cases of lung sarcomatoid carcinoma were analyzed in the present study. Sarcomatoid mesothelioma included 25 cases of pure sarcomatoid growth (pure sarcomatoid mesothelioma) and 6 cases of biphasic mesothelioma showing predominantly sarcomatoid growth. Lung sarcomatoid carcinoma included 5 cases of spindle cell carcinoma and 24 of pleomorphic carcinoma with predominant spindle cell carcinoma component. Minor foci of squamous cell carcinoma and adenocarcinoma component were present in 5 and 19 cases of pleomorphic carcinoma. Carcinosarcoma, giant cell carcinoma and pulmonary blastema were not included in this study.
The anonymized (unlinkable) tissue samples were provided by the Department of Pathology for gene expression analysis and immunohistochemical study. This study is in accordance with the Ethics Guidelines for Human Genome/Gene Research enacted by the Japanese Government for the collection of tissue specimens and was approved by the institutional ethics review committee (Hiroshima University E-48).
Gene Expression Analysis
Formalin-fixed paraffin-embedded sections from six cases of sarcomatoid mesothelioma and six cases of lung sarcomatoid carcinoma were used for gene expression analysis. RNA extraction for gene expression analysis was performed from the spindled tumor cells of these cases. Five 10-μm-thick formalin-fixed paraffin-embedded tissue sections containing >90% spindled tumor tissue were processed for total RNA extraction using the Maxwell 16 LEV RNA FFPE Purification Kit (Promega, Tokyo, Japan) according to the manufacturer’s protocol. After deparaffinization and lysis with proteinase K treatment, the samples were treated with a DNase cocktail for 15 min at room temperature, followed by RNA purification using a MAXWELL 16 instrument according to the manufacturer’s protocol (Promega).
RNA quality was analyzed with an RNA StdSens Analysis kit using an Experion automated electrophoresis system (Bio-Rad Laboratories, Hercules, CA, USA). RNA quantity was estimated with a Qubit RNA HS Kit using a Qubit Fluorometer 2.0 (Molecular Probes/Life Technologies, Carlsbad, CA, USA). The Almac Xcel Array GeneChip (Affymetrix, Santa Clara, CA, USA) contains probe sets of >97 000 transcripts and was used to analyze gene expression profiles. Total RNA was amplified and labeled with a 3′ IVT Labeling Kit (Affymetrix) before hybridization onto the GeneChip. Briefly, 100 ng total RNA was amplified with a SensationPlus FFPE Amplification Kit (Affymetrix) to generate 30 μg of SenseRNA according to the manufacturer’s protocol. Twenty-five micrograms of SenseRNA was labeled with a 3′ IVT Labeling Kit (Affymetrix) and hybridized to a Almac Xcel Array GeneChip (Affymetrix) at 45 °C for 16 h using a GeneChip Hybridization Oven 645 (Affymetrix). The hybridized GeneChip was washed, stained using GeneChip Fluidic Station 450 (Affymetrix) and scanned with a GeneChip Scanner 3000 7G (Affymetrix) using the GeneChip Operating Software (Affymetrix). The data were analyzed using the Gene Expression Console Software (Affymetrix), and further statistical analyses were performed using the Subio Software Platform (Subio, Amami-shi, Japan) to calculate plot graphs, fold change of expression and hierarchical clustering.
Validation of Gene Expression Analysis
The same 12 cases of sarcomatoid mesothelioma and lung sarcomatoid carcinoma that were analyzed for gene expression profiling were used to validate the microarray expression data. The relative mRNA expression of MUC4, a highly expressed gene in lung sarcomatoid carcinoma, and IGF2, highly expressed in sarcomatoid mesothelioma, was assessed with SYBR Green-based real-time RT-PCR using GAPDH as a control. A total of 100 ng RNA was used for mRNA expression with a one-step SYBR Green RT-PCR Kit (Takara-Bio, Tokyo, Japan) using a MX3000P real-time PCR thermal cycler (Stratagene, Agilent Technologies, Tokyo, Japan). The primer pairs used were MUC4-F: CAGGCCACCAACTTCATCG; MUC4-R: ACACGGATTGCGTCGTGAG; IGF2-F: GTGGCATCGTTGAGGAGTG; IGF2-R: CACGTCCCTCTCGGACTTG; GAPDH-F: ACAACTTTGGTATCGTGGAAGG; and GAPDH-R: GCCATCACGCCACAGTTTC. Data analysis was performed using the δδCT method for relative quantification. Briefly, threshold cycles (CT) for GAPDH (reference) and MUC4, IGF2 (samples) were determined in triplicate. The relative expression (rI) was calculated using the formula: rI=2−(CT sample−CT normal).
Immunohistochemistry
Immunohistochemistry was performed using 3-μm tissue sections from the best representative formalin-fixed paraffin-embedded sarcomatoid mesothelioma and lung sarcomatoid carcinoma tissue blocks. All of the immunohistochemical staining was performed with a Benchmark GX automated immunohistochemical station (Ventana, Roche Diagnostics, Tokyo, Japan) using the ultraView Universal DAB Detection Kit (Ventana, Roche Diagnostic, Tokyo, Japan). The antigen retrieval methods and antibodies used in this study are summarized in Table 1. Immunoreactivity was scored as negative (0, no immunostaining) or positive. Positive immunoreactivity was graded as +1 for up to 10% of tumor cells showing positive immunostaining, +2 for >10–50% of the tumor cells, and +3 for >50% of the tumor cells. Only spindled tumor cells from sarcomatoid mesothelioma and lung sarcomatoid carcinoma were evaluated for the immunoreactivity of various markers. Statistical analyses were performed using Fisher’s exact test. Sensitivity, specificity, positive predictive value, negative predictive value and accuracy rate were calculated using a simple 2 × 2 table.
Results
Differential Gene Expression and Validation in Sarcomatoid Mesothelioma and Lung Sarcomatoid Carcinoma
Out of the 97 000 analyzable transcripts on the Almac Xcel Array GeneChip, 2099 statistically significant mRNA transcripts were differentially expressed between sarcomatoid mesothelioma and lung sarcomatoid carcinoma by a more than a two-fold difference (Figure 1, plot graph). The hierarchical clustering of mRNAs with more than a five-fold difference in expression revealed 156 upregulated mRNA transcripts, including IGF2, MEG3, CLIC4 and SPARC, in sarcomatoid mesothelioma and 46 upregulated mRNA transcripts, including MUC4 and Claudin4, in lung sarcomatoid carcinoma (Figure 2, hierarchical clustering; Supplementary Table S1). The mRNA expression hits were validated by real-time RT-PCR of MUC4 and IGF2. MUC4 mRNA expression was negligible in all six sarcomatoid mesothelioma, and the expression was observed in five of the six lung sarcomatoid carcinoma samples. IGF2 mRNA was expressed in all of the sarcomatoid mesothelioma samples, although it was also expressed in three of the six lung sarcomatoid carcinoma samples (detailed data not shown).
The hierarchical clustering of RNA transcripts with more than five-fold differential expression between sarcomatoid mesothelioma and lung sarcomatoid carcinoma revealed 156 upregulated mRNA transcripts, including IGF2, MEG3, CLIC4 and SPARC, in sarcomatoid mesothelioma and 46 upregulated mRNA transcripts, including MUC4 and Claudin4, in lung sarcomatoid carcinoma.
Immunohistochemical Profiles of Sarcomatoid Mesothelioma and Lung Sarcomatoid Carcinoma
The percentage of positivity and immunohistochemical score for MUC4, mesothelioma markers (calretinin, D2-40, WT1) and lung carcinoma markers (TTF-1, p40, Claudin-4) along with the cytokeratins AE1/AE3 and CAM5.2 are shown in Table 2.
MUC4 Expression
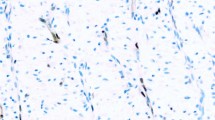
MUC4 expression was observed in the cytoplasm of tumor cells, and the positivity of spindled tumor cells alone was evaluated. MUC4 was also observed in the surrounding normal lung tissue, particularly in bronchial tissue, and was considered an internal positive marker. It was expressed in spindled tumor cells of 21 lung sarcomatoid carcinoma (21/29, 72%; Figure 3b) but none in sarcomatoid mesothelioma (0/31, 0%; Figure 4b). In addition to spindled tumor cells of lung sarcomatoid carcinoma, MUC4 was also expressed in the non-small cell carcinoma component consisting of adenocarcinoma or squamous cell carcinoma in pleomorphic carcinoma. Among lung sarcomatoid carcinoma, 3 cases showed expression in >50% of tumor cells, 9 cases in 10–50% of tumor cells and 9 cases in <10% of tumor cells. Out of the 21 lung sarcomatoid carcinoma cases with MUC4 expression, p40 expression was observed only in 3 cases, TTF-1 in 12 cases and Claudin-4 in 10 cases. Of the nine lung sarcomatoid carcinoma cases without MUC4 expression, p40 expression was observed in three cases, TTF-1 in three cases and Claudin-4 in three cases.
Calretinin, D2-40 and WT1
Calretinin was expressed in the nucleus and cytoplasm of spindled tumor cells of 23 (74%) sarcomatoid mesothelioma and 13 (45%) lung sarcomatoid carcinoma samples, and D2-40 was expressed in the spindled tumor cells of 21 (71%) sarcomatoid mesothelioma and 9 (31%) lung sarcomatoid carcinoma. The immunohistochemical scoring pattern for calretinin expression was not different between sarcomatoid mesothelioma and lung sarcomatoid carcinoma. However, the immunohistochemical scoring pattern for D2-40 expression showed a higher score in sarcomatoid mesothelioma than in lung sarcomatoid carcinoma. WT1 nuclear expression was present in only 6 (19%) sarcomatoid mesothelioma and 1 (3%) lung sarcomatoid carcinoma, revealing it to be a poor immunohistochemical marker to differentiate sarcomatoid mesothelioma from lung sarcomatoid carcinoma.
TTF-1, p40, Claudin-4
Nuclear expression of TTF-1 and P40 was observed in 15 (52%) and 6 (21%) cases of lung sarcomatoid carcinoma, respectively. TTF-1 expression was not observed in sarcomatoid mesothelioma, but p40 expression was observed in 2 (7%) sarcomatoid mesothelioma cases. TTF-1 and/or p40 immunoreactivity was present in 19 of the 29 (66%) cases of lung sarcomatoid carcinoma and 2 of the 31 (7%) cases of sarcomatoid mesothelioma. Claudin-4 and/or TTF-1/p40 immunoreactivity was present in 25 of the 29 (86%) of lung sarcomatoid carcinoma and 2 of the 31 (7%) cases of sarcomatoid mesothelioma. However, p40 expression in sarcomatoid mesothelioma was focal and heterogeneous with an immunohistochemical score of 1.
Cytokeratins, AE1/AE3, CAM5.2
Cytokeratin AE1/AE3 and CAM5.2 expression was present in >90% of both lung sarcomatoid carcinoma and sarcomatoid mesothelioma samples. The majority of sarcomatoid mesothelioma and lung sarcomatoid carcinoma cases showed the expression of both cytokeratins, and the remaining two lung sarcomatoid carcinoma cases and one sarcomatoid mesothelioma case expressed at least one of the two cytokeratins.
Sensitivity and Specificity of Each Marker to Differentially Diagnose Sarcomatoid Mesothelioma and Lung Sarcomatoid Carcinoma
The sensitivity, specificity, positive predictive value, negative predictive value and accuracy rate of each marker differentiating sarcomatoid mesothelioma from lung sarcomatoid carcinoma are shown in Table 3. The negative expression of the carcinoma markers TTF-1 and Claudin-4 showed 100% sensitivity, whereas p40 showed 94%; however, their specificity was restricted around or below 50%. The positive expression of calretinin showed 74% sensitivity and 55% specificity, and D2-40 showed 71% sensitivity and 69% specificity. Although WT1 showed the highest specificity of 97%, its sensitivity was <20%. AE1/AE3 and CAM5.2 showed high 94 and 90% sensitivities and near 0% specificity. In comparison to all of these known immunohistochemical markers, negative expression of MUC4 showed 100% sensitivity and 72% specificity, making the accuracy rate of 87%, the highest among these immunohistochemical markers.
Value of Immunohistochemical Marker Panel to Differentially Diagnosis Sarcomatoid Mesothelioma and Lung Sarcomatoid Carcinoma
MUC4 showed the highest sensitivity and specificity among the immunohistochemical markers for differentiatiation of sarcomatoid mesothelioma from lung sarcomatoid carcinoma. However, the specificity was 72%. Therefore, a combination of various markers was considered. Various combinations of immunohistochemical markers are shown in Table 4. Among the negative immunohistochemical markers, combination of MUC4, TTF-1 and p40 was observed in 26 of the 29 lung sarcomatoid carcinoma cases (90% specificity) and 2 of the 31sarcomatoid mesothelioma cases (93% sensitivity). Combination of MUC4 and Claudin-4 expression was found in 24 of the 29 lung sarcomatoid carcinoma cases (83% specificity) and none of the sarcomatoid mesothelioma cases (100% sensitivity). The combination of MUC4, TTF-1 and Claudin-4 was observed in 26 of the 29 lung sarcomatoid carcinoma cases (90% specificity) and 0 of the 31 sarcomatoid mesothelioma cases (100% sensitivity).
Discussion
Sarcomatoid mesothelioma has the histomorphological feature of spindled tumor cells and resembles many tumors with spindled cells, including true sarcoma or sarcomatoid carcinomas. The immunohistochemical reactivity to cytokeratin remains critical to differentiate it from true sarcomas. However, differentiating sarcomatoid mesothelioma from lung sarcomatoid carcinoma is challenging, as the histomorphological and immunohistochemical characteristics are extremely similar. For this reason, clinical and/or gross evidence of an extrapulmonary location is indispensable for its diagnosis. Although the mesothelioma markers calretinin and D2-40 have been utilized to differentiate sarcomatoid mesothelioma from lung sarcomatoid carcinoma, they are not absolute, as their sensitivity and specificity are not sufficiently high. Although we previously reported the sensitivity of calretinin (78%) and D2-40 (87%), specificity was not high for calretinin (41%) and D2-40 (74%).7 Our past and present data on calretinin and D2-40 were similar to reports by Ordonez et al4and Padgett et al.6 Considering the low specificity of calretinin, D2-40 is considered the single most important immunohistochemical marker for its differentiation. However, in our practical experience, it is still very difficult to interpret the reactivity of D2-40 in these tumors, particularly in cases showing prominent fibro-collagenous proliferation.
TTF-1, a lung adenocarcinoma marker, and p40, a squamous cell carcinoma marker, have emerged as useful markers for non-small cell lung carcinoma10, 11 and are thus supposed to be expressed in pleomorphic lung carcinoma. TTF-1 might be identified as a novel marker differentiating pleomorphic carcinoma from sarcomatoid mesothelioma because of its low expression in sarcomatoid mesothelioma. However, in this study, despite their specificity of 100 or 94%, the sensitivity of TTF-1 (51%) and p40 (21%) are not good to distinguish sarcomatoid mesothelioma and lung sarcomatoid carcinoma. Though p40 expression is good marker of squamous cell carcinoma, it has been also reported in a few mesothelioma cases.12 In this study too, we observed p40 expression in two sarcomatoid mesothelioma cases but very focal and heterogeneous, unlike its expression in squamous cell carcinoma. Claudin-4, which is reported to be a very reliable universal carcinoma marker differentiating epithelioid mesothelioma from various carcinomas,13, 14 showed limited value in lung sarcomatoid carcinoma cases. In this study, only half of lung sarcomatoid carcinoma expressed Claudin-4, and its punctate expression in the cytoplasm of spindled cells of lung sarcomatoid carcinoma resembled that of the punctate expression in the cytoplasm of sarcomatoid mesothelioma. TTF-1, p40 and Claudin-4 expression can be reliable markers for pleomorphic carcinomas with a prominent carcinoma component, such as adenocarcinoma or squamous cell carcinoma.
In this study, we analyzed all of the genes expressed in sarcomatoid mesothelioma and lung sarcomatoid carcinoma with the aim of identifying novel markers for their differential diagnosis. Although frozen tissue yields better and less degradable RNA for gene expression analysis, we preferred formalin-fixed paraffin-embedded tissue samples because they included the microscopically identifiable spindle cell tumor tissue. For this analysis, we have to amplify the small amount of RNA extracted from the formalin-fixed paraffin-embedded tissue before hybridization to the GeneChip. The Almac Xcel GeneChip from Affymetrix, which we used here, has been reported to produce identical results to the GeneChip using RNA derived from frozen tissue samples. In addition, it contains proprietary Almac-sequenced data and filtered public data for biomarker discovery and the validation of oncogene-related transcripts for a much higher detection rate in degraded samples.
From the differential expression analysis, a more than five-fold expression change in IGF2, CLIC4 and SPARC was observed in sarcomatoid mesothelioma, and IGF2 expression was validated by real-time RT-PCR. We did not uncover significant differential expression of IGF2 between sarcomatoid mesothelioma and lung sarcomatoid carcinoma (data not shown). The discrepancy between the microarray data and real-time RT-PCR data can be explained because IGF2 mRNA expression on a microarray chip is the relative expression between both lung sarcomatoid carcinoma and sarcomatoid mesothelioma but in a different quantity. We later investigated the immunohistochemical expression IGF2, CLIC4 and SPARC proteins in sarcomatoid mesothelioma and lung sarcomatoid carcinoma. However, there was no significant differential expression of these proteins between lung sarcomatoid carcinoma and sarcomatoid mesothelioma, limiting their applicability as an immunohistochemical positive marker of sarcomatoid mesothelioma.
In contrast, microarray gene expression analysis showed increased expression of MUC4 in lung sarcomatoid carcinoma compared with that of sarcomatoid mesothelioma, and we found negligible MUC4 mRNA expression in sarcomatoid mesothelioma at the mRNA level. MUC4 stands for member of mucin protein of high molecular weight glycoprotein.15 It is expressed in various normal epithelium of the respiratory tract, particularly in the trachea and bronchi16 and in the epithelium of the digestive and urogenital tracts.17 MUC4 expression has been reported in various human carcinomas, including pancreatic,18 breast19 and lung adenocarcinoma.20 Llinares et al21 reported the diagnostic value of MUC4 expression in distinguishing epithelioid mesothelioma and lung adenocarcinoma. They found that MUC4 was expressed in 0 of the 41 epithelioid mesotheliomas and in 32 of the 35 (91%) lung adenocarcinoma. To our knowledge, this report has not been validated by other laboratories, as the antibody to MUC4 was not commercially available in the past. We observed MUC4 expression in lung adenocarcinoma and lung squamous cell carcinoma and observed no expression in epithelioid mesothelioma using a commercially available anti-MUC4 antibody. The current study is the first report to describe MUC4 expression in lung sarcomatoid carcinoma and no MUC4 expression in sarcomatoid mesothelioma. We observed a high specificity (72%) and absolute sensitivity (100%) for negative MUC4 expression to differentiate sarcomatoid mesothelioma from lung sarcomatoid carcinoma, with an accuracy rate of 87%. These values are far better than any previously identified immunohistochemical markers differentiating sarcomatoid mesothelioma from lung sarcomatoid carcinoma.
The sensitivity of MUC4 expression as a negative marker was the highest of the immunohistochemical markers in this study. Lung sarcomatoid carcinoma cases showing MUC4 expression (21 cases) also demonstrated co-expression of TTF-1 in 12 cases, Claudin-4 in 10 cases and p40 in 3 cases. Furthermore, lung sarcomatoid carcinoma cases without MUC4 expression showed TTF-1 expression in three cases, p40 in three cases and Claudin-4 in three cases. Therefore, MUC4 expression has better additional value of the immunohistochemical markers for the differential diagnosis of sarcomatoid mesothelioma from lung sarcomatoid carcinoma. The sensitivity of these markers can be improved by combining two or more, and the addition of TTF-1 and Claudin-4 to MUC4 expression improved the accuracy rate up to 95% for the differential diagnosis of sarcomatoid mesothelioma from lung sarcomatoid carcinoma.
In conclusion, we identified a novel immunohistochemical marker MUC4 that differentiates sarcomatoid mesothelioma from lung sarcomatoid carcinoma by applying whole gene expression analysis. The combination of MUC4 with TTF-1/p40 and Claudin-4 improved the sensitivity and specificity for differential diagnosis. Therefore, we propose including MUC4 as an additional negative marker to the immunohistochemical marker panel to differentiate sarcomatoid mesothelioma from lung sarcomatoid carcinoma.
References
Delgermaa V, Takahashi K, Park EK et al. Global mesothelioma deaths reported to the World Health Organization between 1994 and 2008. Bull World Health Organ 2011;89:716–724, 24A–24C.
Roggli V, Churg A, Chirieac LR et al, Sarcomatoid, desmoplastic, and biphasic mesothelioma. In: Travis WD, Brambilla E, Burke S et al, (eds). WHO Classification of Tumours of the Lung, Pleura, Thymus, and Heart. IARC: Lyon, France, 2015, pp 165–168.
Husain AN, Colby T, Ordonez N et al. Guidelines for pathologic diagnosis of malignant mesothelioma: 2012 update of the consensus statement from the International Mesothelioma Interest Group. Arch Pathol Lab Med 2013;137:647–667.
Ordonez NG . D2-40 and podoplanin are highly specific and sensitive immunohistochemical markers of epithelioid malignant mesothelioma. Hum Pathol 2005;36:372–380.
Chu AY, Litzky LA, Pasha TL et al. Utility of D2-40, a novel mesothelial marker, in the diagnosis of malignant mesothelioma. Mod Pathol 2005;18:105–110.
Padgett DM, Cathro HP, Wick MR et al. Podoplanin is a better immunohistochemical marker for sarcomatoid mesothelioma than calretinin. Am J Surg Pathol 2008;32:123–127.
Takeshima Y, Amatya VJ, Kushitani K et al. Value of immunohistochemistry in the differential diagnosis of pleural sarcomatoid mesothelioma from lung sarcomatoid carcinoma. Histopathology 2009;54:667–676.
Kushitani K, Amatya VJ, Mawas AS et al. Use of anti-Noxa antibody for differential diagnosis between epithelioid mesothelioma and reactive mesothelial hyperplasia. Pathobiology 2016;83:33–40.
Kerr KM, Pelosi G, Austin JHM et al, Pleomorphic, spindle cell, and giant cell carcinoma. In: Travis WD, Brambilla E, Burke AP et al, (eds). WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart, 4th edn. IARC: Lyon, France, 2015, pp 88–90.
Travis WD, Noguchi M, Yatabe Y et al, Adenocarcinomal. In: Travis WD, Brambilla E, Burke AP et al, (eds). WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart, 4th edn. IARC: Lyon, France, 2015, pp 26–37.
Tsao M-S, Brambilla E, Nicholson AG et al, Squamous cell carcinoma. In: Travis WD, Brambilla E, Burke AP et al, (eds). WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart, 4th edn. IARC: Lyon, France, 2015, pp 51–55.
Tatsumori T, Tsuta K, Masai K et al. p40 is the best marker for diagnosing pulmonary squamous cell carcinoma: comparison with p63, cytokeratin 5/6, desmocollin-3, and sox2. Appl Immunohistochem Mol Morphol 2014;22:377–382.
Ordonez NG . Value of claudin-4 immunostaining in the diagnosis of mesothelioma. Am J Clin Pathol 2013;139:611–619.
Facchetti F, Lonardi S, Gentili F et al. Claudin 4 identifies a wide spectrum of epithelial neoplasms and represents a very useful marker for carcinoma versus mesothelioma diagnosis in pleural and peritoneal biopsies and effusions. Virchows Arch 2007;451:669–680.
Chaturvedi P, Singh AP, Batra SK . Structure, evolution, and biology of the MUC4 mucin. FASEB J 2008;22:966–981.
Copin MC, Devisme L, Buisine MP et al. From normal respiratory mucosa to epidermoid carcinoma: expression of human mucin genes. Int J Cancer 2000;86:162–168.
Audie JP, Janin A, Porchet N et al. Expression of human mucin genes in respiratory, digestive, and reproductive tracts ascertained by in situ hybridization. J Histochem Cytochem 1993;41:1479–1485.
Moniaux N, Chaturvedi P, Varshney GC et al. Human MUC4 mucin induces ultra-structural changes and tumorigenicity in pancreatic cancer cells. Br J Cancer 2007;97:345–357.
Rakha EA, Boyce RW, Abd El-Rehim D et al. Expression of mucins (MUC1, MUC2, MUC3, MUC4, MUC5AC and MUC6) and their prognostic significance in human breast cancer. Mod Pathol 2005;18:1295–1304.
Kwon KY, Ro JY, Singhal N et al. MUC4 expression in non-small cell lung carcinomas: relationship to tumor histology and patient survival. Arch Pathol Lab Med 2007;131:593–598.
Llinares K, Escande F, Aubert S et al. Diagnostic value of MUC4 immunostaining in distinguishing epithelial mesothelioma and lung adenocarcinoma. Mod Pathol 2004;17:150–157.
Acknowledgements
We thank Ms Naomi Fukuhara for her administrative assistance. This work was supported in part by the Japan Society for the Promotion of Science, Grants-in-Aid Scientific Research No. JP26460452 (YT) from the Ministry of Education, Science, Sports and Culture. Part of this study was performed at the Analysis Center of Life Science, Hiroshima University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Amatya, V., Kushitani, K., Mawas, A. et al. MUC4, a novel immunohistochemical marker identified by gene expression profiling, differentiates pleural sarcomatoid mesothelioma from lung sarcomatoid carcinoma. Mod Pathol 30, 672–681 (2017). https://doi.org/10.1038/modpathol.2016.181
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2016.181
This article is cited by
-
Monoclonal Caveolin 1 Expression in the Differential Diagnosis of Malignant Pleural Mesothelioma and Pulmonary Adenocarcinoma: Is it Useful?
Pathology & Oncology Research (2020)
-
Sarcoma with MGA–NUTM1 fusion in the lung: an emerging entity
Virchows Archiv (2020)
-
Reply to ‘MUC4 staining in sarcomatoid carcinomas’ by Berg et al.
Modern Pathology (2019)
-
MUC4 Staining in Sarcomatoid Carcinomas
Modern Pathology (2019)
-
MUC4 immunohistochemistry is useful in distinguishing epithelioid mesothelioma from adenocarcinoma and squamous cell carcinoma of the lung
Scientific Reports (2018)