Abstract
By analysing ancient DNA (aDNA) from 74 14C-dated individuals of the extinct South Island giant moa (Dinornis robustus) of New Zealand, we identified four dyads of closely related adult females. Although our total sample included bones from four fossil deposits located within a 10 km radius, these eight individuals had all been excavated from the same locality. Indications of kinship were based on high pairwise genetic relatedness (rXY) in six microsatellite markers genotyped from aDNA, coupled with overlapping radiocarbon ages. The observed rXY values in the four dyads exceeded a conservative cutoff value for potential relatives obtained from simulated data. In three of the four dyads, the kinship was further supported by observing shared and rare mitochondrial haplotypes. Simulations demonstrated that the proportion of observed dyads above the cutoff value was at least 20 times higher than expected in a randomly mating population with temporal sampling, also when introducing population structure in the simulations. We conclude that the results must reflect social structure in the moa population and we discuss the implications for future aDNA research.
Similar content being viewed by others
Introduction
Because of the challenges in characterising nuclear DNA from degraded substrates, microsatellite-based analyses such as that of genetic structuring (Pritchard et al., 2000; Hardy and Vekemans, 2002), migration rates (Beerli and Felsenstein, 1999; Wilson and Rannala, 2003) and kinship and relatedness (Queller et al., 1993; Kalinowski et al., 2006) have been largely absent from ancient DNA (aDNA) research. However, with new methods available for identifying microsatellites markers from aDNA (Allentoft et al., 2009) and the development of strict protocols for the subsequent data generation (Taberlet et al., 1996; Allentoft et al., 2011), microsatellite analyses of extinct taxa are no longer beyond reach (Ishida et al, 2012; Nyström et al, 2012; Allentoft et al, 2014). Here we employ microsatellite DNA profiling on a large assemblage of radiocarbon-dated fossils of the extinct New Zealand moa (Aves: Dinornithiformes) in an attempt to gain new insights into the palaeobiology of this extinct megafauna.
Nine species of moa are currently recognised (Bunce et al, 2009), all of which were large browsing ratites endemic to New Zealand. They became extinct as a result of overhunting and habitat destruction shortly after Polynesians settled in the thirteenth century (Anderson, 1989; Holdaway and Jacomb, 2000; Allentoft et al., 2014; Holdaway et al., 2014). As is the case for most extinct taxa, little is known about their biology except some insights into their feeding habits, gained from their gut contents and genetic and morphological analyses of coprolites (e.g., Burrows et al., 1981; Burrows, 1989; Wood et al., 2008, 2013). It has also been demonstrated that moa displayed reverse sexual dimorphism with the females being up to 280% heavier than the males (Bunce et al., 2003; Huynen et al., 2003). Moreover, Allentoft et al. (2010) demonstrated by a molecular sexing technique that there was an overall 5:1 excess of females among 267 individuals sampled in North Canterbury. In particular, moa fossils from the shallow lake at Pyramid Valley, excavated periodically since 1939, displayed an extreme excess of 49 females to 2 males of South Island giant moa (Dinornis robustus). Hypotheses involving female territoriality and seasonal egg incubation by males (both observed among extant ratites) have been invoked to explain the absence of males from some deposits. From the observation of an even sex ratio among juvenile moa of the taxon Euryapteryx curtus (Allentoft et al., 2010), but not among the adults, a higher mortality among males has also been suggested. Incubation by males has been suggested by Huynen et al. (2010) who identified putative moa male DNA on the surface of their eggshell.
Although these are just fragmentary insights into moa biology, they still place moa among the best studied extinct animals (summarised in Worthy and Holdaway, 2002 and Allentoft and Rawlence, 2012), and promote them as promising ‘model organisms’ for research into the social structure and behaviour of extinct species. In an attempt to obtain a deeper understanding of moa biology, and explore the potential in using nuclear genetic data from fossils, we re-analysed a recently published aDNA data set (Allentoft et al., 2014). This data set included both mitochondrial DNA (mtDNA) sequences and six genotyped microsatellite loci in 74 radiocarbon-dated individuals of D. robustus, excavated from four sites within a 10 km radius in North Canterbury, South Island (Figure 1), and covering ~4200 years of the mid-to-late Holocene. D. robustus was the largest of the moa species with females weighing up to 250 kg (Worthy and Holdaway, 2002; Bunce et al., 2003; Brassey et al., 2013). We employed a range of methods to explore the genetic relatedness among the 74 individuals. The application of microsatellite profiling to fossil assemblages is interesting from both biological and methodological perspectives. If such ‘DNA fingerprinting’ technology can be successfully applied to extinct species, it could result in significant insights into their biology and behaviour, taking population palaeogenetics to a new level.
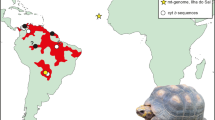
Sampling localities. Map of New Zealand’s South Island with the sampling area in North Canterbury indicated. This study includes genetic data from four proximate fossil sites but all four identified dyads of related Dinornis robustus females were from Pyramid Valley (lower image), which has been excavated periodically since the 1930s. The exact locations at Pyramid Valley are shown in Supplementary Figure S4.
Materials and Methods
Data generation
The microsatellite, mtDNA, and radiocarbon age data analysed in this study were first presented in Allentoft et al. (2014), where details of sampling, DNA extraction, genotyping validation and radiocarbon dating are given. The D. robustus data set includes mtDNA control region sequences from 87 radiocarbon dated individuals and multilocus microsatellite profiles from 74 of these. The 74 samples are from four fossil deposits (Pyramid Valley; Bell Hill Vineyard; Glencrieff; Glenmark) within a 10 km radius in North Canterbury, South Island, New Zealand (Figure 1).
Briefly, left tibiotarsi of moa were sampled by ‘coring’ at mid-shaft: 200 mg of bone material was powdered and incubated overnight in a buffer (EDTA, proteinase K, dithiothreitol and Triton-X). The DNA was then isolated and purified using Qiagen (Valencia, CA, USA) silica spin columns. A mtDNA control region sequence of 340 bp (excluding primers) was amplified from 87 individuals using two sets of overlapping primers (Bunce et al., 2003). The associated GenBank accession numbers for the individual mtDNA sequences are presented in Allentoft et al. (2014). Six moa microsatellite markers were amplified successfully in 74 of the 87 individuals using rigorous methods for generating high-quality microsatellite data from ancient DNA as outlined previously (Allentoft et al., 2011).
ML-RELATE
Two methods were used to identify relatives in the data: (i) a maximum likelihood-based approach, as implemented in the software ML-RELATE (Kalinowski et al., 2006) and (ii) a more qualitative ad hoc assessment of observed and simulated relatedness indices (rXY), combined with radiocarbon dates and mtDNA control region haplotypes.
ML-RELATE applies a maximum likelihood method based on relatedness values to estimate if each pair of individuals (dyads) have a higher probability of being unrelated, full siblings, half siblings or parent–offspring (Kalinowski et al., 2006). The confidence intervals for all pairwise relationships among the 74 individuals were assessed according to a 95% significance level and 10 000 permutations.
Simulating relatives
In the second method, pairwise rXY values (Queller and Goodnight (1989), adapted by Oliehoek et al. (2006)) were calculated between all dyads using the R package DEMERELATE (Kraemer and Gerlach, 2013). Closely related moa individuals should show high allele sharing and similar geological ages (calibrated 14C ages), because each passing generation of random mating reduces the relatedness of individuals on the same genealogical branch by half. To examine this relationship we plotted the rXY value for each dyad against the number of years separating the two individuals.
To assess the significance of the rXY values in our observed data, we simulated 10 000 random offspring dyads of each of the relatedness classes ‘non-related’, ‘half siblings’, and ‘full siblings’ and calculated the rXY distribution in each class (Supplementary Figure S1). The 95% quantile for non-related individuals was 0.49, meaning that 95% of the dyads with no close family relations displayed rXY values below 0.49. The corresponding 95% values for simulated dyads of half siblings and full siblings were 0.68 and 0.90, respectively (Supplementary Figure S1). We therefore conservatively selected an rXY of 0.68 as a cutoff value for considering dyads in our empirical data set as potential relatives.
Temporal sampling simulations
As it is not known how the temporal structure of the sampling could influence the distribution of relatedness, we performed an additional series of simulations. These were conducted using FASTSIMCOAL2 (Excoffier et al, 2013) to emulate the empirical distribution of sampling times (that is, median-calibrated radiocarbon ages) from the moa population. We simulated data from six microsatellite loci under a random mating population with a constant effective population size (Ne) of 2000 individuals, which yielded an expected heterozygosity (HE) and number of alleles very similar to the observed ones. This Ne was chosen based on opportunistic trial and error, as there is to our knowledge no way of analytically deriving the equilibrium theta under a generalised microsatellite mutation model (Fu and Chakraborty, 1998) such as that implemented in FASTSIMCOAL2. Note that these simulations were done using a constant-sized population at mutation-drift equilibrium, hence theta deviates slightly from that inferred in Allentoft et al. (2014) where a flexible demographic model was used. We performed 100 replicates of this simulation and calculated rXY values as above. To compare the empirical rXY values with this temporal null distribution of simulated relatedness, we calculated how many of the pseudo-observed dyads (99 × 2701 dyads in total) that showed an rXY >0.68. Moreover, we calculated how often this simulation would produce dyads with rXY >0.68 that were separated by 50 years or less (dyads50 in the following), which could be argued as the realistic span of temporal separation for parent–offspring or siblings. Hence we could assess the significance of observing such dyads in our empirical data. We also plotted the distribution of rXY values among the dyads50 only.
Spatiotemporal sampling simulations
The presence of population subdivision (that is, spatial structure) in D. robustus could lead to elevated rXY values in our spatially restricted sampling because we would tend to sample relatives more often than in a random mating, unstructured population of the same effective size. To assess the combined impact of population subdivision and temporal sampling, we performed another series of simulations in FASTSIMCOAL2 where we introduced an island-model type of structure in addition to the temporal sampling described above. We assumed ten demes each of Ne=200, connected by three different levels of migration: high (m=0.05), medium (m=0.005) and low (m=0.0005). We sampled just one of the ten demes to mimic spatially restricted sampling in a structured population. For each scenario we performed 100 replicate simulations and calculated the pairwise rXY among all dyads in each replication.
mtDNA
We assessed the mtDNA similarity of the proposed relatives. Different mtDNA profiles would exclude a number of possible family relationships such as full siblings, half siblings related through the mother and mother–offspring. Conversely, identical haplotypes support kinship, but the strength of this as evidence depends on the frequency of the given haplotype in the population. On the basis of the 87 available mtDNA profiles (Allentoft et al., 2014), we could estimate the haplotype frequencies in the D. robustus population and use this information in our evaluation. To visualise the genetic structure and diversity of the mtDNA sequences we generated a median-joining haplotype network with NETWORK v.4.5 (Fluxus-engineering.com; Bandelt et al, 1999).
Results
Detailed descriptions of the genetic diversity and the radiocarbon ages have been presented previously (Allentoft et al., 2012a, 2012b, 2014) and will be mentioned only briefly here. The total data set encompassed 74 individuals genotyped in six microsatellite markers, except for four individuals that could only be genotyped in five of the markers despite several attempts. The median calibrated radiocarbon ages of the 74 individuals profiled for microsatellites ranged from 4837 to 602 years before present (Allentoft et al, 2014).
Deviation from Hardy–Weinberg proportions (α=0.05) was observed in one marker but not after Bonferroni correction and no tests for linkage disequilibrium among loci were significant (Allentoft et al., 2014). Moreover, we did not find evidence of scoring bias from null alleles, allelic drop-out, or stuttering in the data (Allentoft et al., 2014). The Holocene D. robustus population of North Canterbury displayed an expected microsatellite heterozygosity (HE) of 0.72 (see allele frequencies in Supplementary Table S1) and had 29 haplotypes in the targeted mtDNA control region (Allentoft et al., 2014).
ML-RELATE identified a surprisingly high number of potential relatives in the sample. Of 2701 pairwise relationships tested among the 74 individuals, ML-RELATE estimated 551 family relations (71 full siblings, 344 half siblings and 136 parent–offspring relationships). The number seems unrealistically high, given the >4000 year timeframe covered by the data and the fact that many of these putative relatives were separated by hundreds of years. The average age separation between identified full siblings in ML-RELATE was 988 years, whereas the same numbers were 1019 years and 1021 years for parent–offspring and unrelated individuals, respectively. Although the numbers indicated a weak tendency towards a smaller average age separation in related individuals, it was not significant (t-test, P=0.50). We conclude that ML-RELATE overestimated the number of family relations in the data, which is likely the result of the limited resolution power of the six microsatellite markers. When excluding all pairwise relations where the confidence sets included a status of ‘unrelated’, 113 dyads remained.
The simulation cutoff approach was more conservative in inferring possible relatives. Of the 2701 pairwise combinations (dyads) among the 74 individuals in our observed data set, only 13 had rXY values exceeding the 0.68 cutoff for relatives obtained through simulations (Figure 2). Again, some of these putative relatives were separated by centuries or millennia (Figure 2), and assuming that the radiocarbon dates have normal levels of accuracy, these could not have been related and therefore constitue false positives. Four of the dyads (0.15%), however, clustered out with both high rXY values (0.78–0.88) and similar calibrated radiocarbon ages (Figure 2, Table 1). Among the 124 dyads in which the two individuals were separated by <50 years of sampling time (dyads50; Figure 2), these four dyads with high rXY constitute 3.2%, which is ~22 times higher frequency than found across all dyads. The eight individuals involved were all adult females excavated from the same area of the Pyramid Valley site (Figure 1,Table 1, Supplementary Figure S4).
Relatedness. (a) Relatedness (rXY) values between all 74 individuals (2701 dyads) plotted against their difference in radiocarbon age. The 95% upper boundary for simulated half siblings (rXY=0.68) was used as cutoff for potential relatives. An age difference of 0–50 years is highlighted as it represents the realistic temporal gap between related individuals (parent–offspring or siblings). Four dyads containing putative relatives were identified with an rXY >0.68 and within the 50-year-separation window. (b–d) rXY Distributions for simulated spatio-temporal data sets based on three different migration rates.
In the temporally simulated data, emulating a randomly mating population sampled through time, we observed 554 (0.2%) of 270 100 dyads across the 100 simulation runs that had rXY values exceeding the 0.68 cutoff. However, only 21 (0.008%) of these were also dyads50 and hence qualifying as potential relatives according to our criteria. This is ~19 times less than observed in the empirical data (0.15%). Equivalently, given the frequency of putative relatives in the simulated null distribution (0.008%), the binomial probability of observing four or more dyads of relatives among the 2701 observed dyads is just 0.008%. Hence our observed data are highly unlikely to have been drawn from a null distribution representing random mating. When focusing on the rXY distribution among the 12 400 simulated dyads50 there were only 21 (0.16%) with rXY >0.68, which is 20 times less than observed among our empirical dyads50 (3.2%) (Supplementary Figure S2). Moreover, none of the 100 replicate simulation runs had more than two dyads50 with rXY >0.68, and >80% of the simulations had zero (Supplementary Figure S3). Again, this is in contrast to the four such dyads observed in the empirical data (Figure 2).
The spatiotemporal simulations did not produce more relatives according to the threshold criteria (Figure 2). In addition, the overall distribution of pairwise rXY values differed considerably from our observed distribution (Figure 2). When structure was introduced in the simulations there was a clear negative correlation between rXY and the temporal separation between two individuals in the sampled subpopulation. This is not surprising as allele sharing at any point in time is higher within than among demes in structured populations. This genetic similarity will, however, decrease with temporal separation because of genetic drift and because incoming migrants are likely to contribute new alleles from demes with different allelic frequencies.
mtDNA
In three of the four putatively related dyads, the two individuals shared the same mitochondrial haplotype. The frequencies of these haplotypes in the North Canterbury population of D. robustus were 0.046 (A), 0.047 (B) and 0.027 (C) (Figure 3, Table 1). The probability of randomly picking the same haplotype in two individuals of a panmictic population is equal to the haplotype frequency. Even allowing for sampling uncertainty in the estimate of the haplotype frequencies, the low frequencies of the shared mtDNA haplotypes in dyads A, B and C strongly support their family relationships. The two individuals in dyad D had different haplotypes that could be explained by the two individuals being related only through the father.
mtDNA haplotype network. Haplotype network based on ∼340 bp of mtDNA control region from 87 D. robustus individuals. The colour composition is defined by the locality where each specimen was found. A, B and C mark the three rare haplotypes that are shared respectively within three of the four dyads with relatives (see Table 1 for details). Open circles reflect single mutations separating the haplotypes.
Discussion
By combining three different sources of information represented by microsatellites, mtDNA and radiocarbon ages, we were able to identify related individuals among fossil moa bones that are thousands of years old. However, the resolution power of six microsatellite markers can be limited when determining relatedness. This is evident from the wide distribution of rXY values in each category of simulated relatives (Supplementary Figure S1), and therefore we did not attempt to determine the exact degree of kinship for the putative relatives. Regardless, our temporal simulations demonstrated that related females were in excess in our empirical data set compared with a population with random mating, and we were able to reject population structure and the nature of our sampling as a potential driver for this observation. We therefore conclude that moa individuals of different social and relatedness classes were not randomly distributed in the landscape and that some behavioural aspect of moa biology must account for these results.
Other observations support this notion. For example, all four putative related dyads consisted of adult females that died in close proximity at Pyramid Valley (Table 1, Supplementary Figure S4). Although 60% of the samples were from females from Pyramid Valley, the naive probability of all four related dyads belonging to this class of individuals is only 1.5%. Also, Allentoft et al. (2010) showed a significant difference in the distribution of sexes and juveniles from the proximate fossil sites at Pyramid Valley and Bell Hill Vineyard (Figure 1). At Pyramid Valley, only two of 49 D. robustus individuals sampled were identified genetically as males and this extreme skew is likely a combination of higher mortality among males combined with behavioural differences or differential habitat preferences between the sexes (Allentoft et al., 2010). The social behaviour in extant ratites varies greatly between taxa but in southern cassowaries (Casuarius casuarius), for example, males and females have separate territories (Moore, 2007), except for a short period when breeding. Ostriches (Struthio camelus) often form flocks with many females (related or not) and only one male. If adult female moa from each cohort were particularly attracted to the habitat around Pyramid Valley, whether foraging in groups of females or alone, it could explain the non-random relatedness distribution in our sample. It has previously been suggested that moa females maintained home ranges around the lakes, whereas males may have been more mobile (Allentoft et al., 2010).
The social structure reflected in our data could not have been uncovered with mtDNA data alone. However, nuclear genetic data are challenging to obtain from ancient samples and for microsatellites there are fundamental issues with allelic dropout that need to be carefully assessed when working with degraded DNA (Navidi et al., 1992; Taberlet et al., 1996; Miller et al., 2002; Roeder et al., 2009; Allentoft et al., 2011). In addition, most of the methods developed to analyse microsatellite markers are not readily compatible with ancient data, which often represent individuals who lived at different points in time. Hence, our study has exploratory value in evaluating the kind of biological information that can be extracted from a highly heterochronous microsatellite data sets. Whereas the ML-RELATE method identified many false-positive relatives, the data simulation approach provided a more sensible null distribution of rXY values for comparison with the observed distribution. This is likely because the simulation framework offers a possibility to mimic temporal sampling, which is crucial when the samples are fossils spanning several millennia. This also explains why data simulation approaches such as approximate Bayesian computation (ABC) are being increasingly applied in aDNA studies when past population dynamics are under scrutiny (Anderson et al., 2005; Chan et al., 2006; Bramanti et al., 2009; Ramakrishnan and Hadly., 2009; Ghirotto et al., 2010; Gamba et al., 2012; Allentoft et al., 2014).
We have here shown that data simulations can be used not only to recover large scale ancient population dynamics from aDNA data but also to investigate patterns of social structure and kinship in past populations. This implies in principle that palaeobiological research can explore an area of population genetics that has, until now, been restricted to extant species. However, it is clear that large assemblages of fossils with excellent biomolecule preservation are rare, and the comprehensive moa fossil record in New Zealand is arguably unique in terms of chronological precision (Holdaway et al., 2014). But the methods we present are not only applicable to large assemblages of extinct megafauna with hundreds of associated radiocarbon dates. Microsatellites are often applied to measure changes in genetic diversity across centuries or decades in extant species, based on museum collections (Groombridge et al., 2000; Whitehouse and Harley, 2001; Johnson et al, 2004; Harper et al, 2006; Petren et al, 2010; Bourke et al, 2010; White et al, 2014) and our simulation approach should be equally suitable to address question of kinship or social structure in such data sets. Our methodological framework could also be applied in association with aDNA recovered from human burials where the chronology is usually well established and where questions related to social structure and kinship are often of great interest to the archaeological community.
Data archiving
As noted in the text, radiocarbon dates and Genbank accession numbers for all mtDNA sequences are provided elsewhere (Allentoft et al., 2014). Microsatellite allele frequences are provided in the online Supplementary Table S1 and individual allele calls are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.hf876.
References
Allentoft ME, Bunce M, Scofield RP, Hale ML, Holdaway RN . (2010). Highly skewed sex ratios and biased fossil deposition of moa: ancient DNA provides new insight on New Zealand's extinct megafauna. Quat Sci Rev 29: 753–762.
Allentoft ME, Collins M, Harker D, Haile J, Oskam CL, Hale ML et al. (2012a). The half-life of DNA in bone: measuring decay kinetics in 158 dated fossils. Proc Royal Soc B: Biol Sci 279: 4724–4733.
Allentoft ME, Heller R, Oskam CL, Lorenzen ED, Hale ML, Gilbert MTP et al. (2014). Extinct New Zealand megafauna were not in decline before human colonization. Proc Natl Acad Sci USA 111: 4922–4927.
Allentoft ME, Oskam C, Houston J, Hale ML, Gilbert MTP, Rasmussen M et al. (2011). Profiling the dead: generating microsatellite data from fossil bones of extinct megafauna—protocols, problems, and prospects. PLoS One 6: e16670.
Allentoft ME, Rawlence NJ . (2012). Moa's Ark or volant ghosts of Gondwana? Insights from nineteen years of ancient DNA research on the extinct moa (Aves: Dinornithiformes) of New Zealand. Ann Anat 194: 36–51.
Allentoft ME, Schuster S, Holdaway RN, Hale ML, McLay E, Oskam C et al. (2009). Identification of microsatellites from an extinct moa species using high-throughput sequence data. Biotechniques 46: 195–200.
Allentoft ME, Scofield RP, Oskam CL, Hale ML, Holdaway RN, Bunce M . (2012b). A molecular characterisation of a newly discovered megafaunal fossil site in North Canterbury, South Island, New Zealand. J Royal Soc of New Zealand 42: 241–256 doi:10.1080/03036758.2011.574821.
Anderson A . (1989) Prodigious Birds. Cambridge University Press: Cambridge, England: Cambridge, England.
Anderson CNK, Ramakrishnan U, Chan YL, Hadly EA . (2005). Serial SimCoal: a population genetics model for data from multiple populations and points in time. Bioinformatics 21: 1733–1734.
Bandelt HJ, Forster P, Röhl A . (1999). Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16: 37–48.
Beerli P, Felsenstein J . (1999). Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics 152: 763–773.
Bourke B, Frantz A, Lavers C, Davison A, Dawson D, Burke T . (2010). Genetic signatures of population change in the British golden eagle (Aquila chrysaetos. Conservation Genetics 11: 1837–1846 doi:10.1007/s10592-010-0076-x.
Bramanti B, Thomas MG, Haak W, Unterlaender M, Jores P, Tambets K et al. (2009). Genetic discontinuity between local hunter-gatherers and central europe's first farmers. Science 326: 137–140.
Brassey CA, Holdaway RN, Packham AG, Anné J, Manning PL, Sellers WI . (2013). More than one way of being a moa: differences in leg bone robustness map divergent evolutionary trajectories in Dinornithidae and Emeidae (Dinornithiformes). PLoS One 8: e82668.
Bunce M, Worthy TH, Ford T, Hoppitt W, Willerslev E, Drummond A et al. (2003). Extreme reversed sexual size dimorphism in the extinct New Zealand moa Dinornis. Nature 425: 172–175.
Bunce M, Worthy TH, Phillips MJ, Holdaway RN, Willerslev E, Haile J et al. (2009). The evolutionary history of the extinct ratite moa and New Zealand Neogene paleogeography. Proc Natl Acad Sci USA 106: 20646–20651.
Burrows CJ . (1989). Moa browsing: evidence from the Pyramid Valley mire. N Z J Ecol 12: 51–56.
Burrows CJ, McCulloch B, Trotter MM . (1981). The diet of moas based on gizzard contents samples from Pyramid Valley, North Canterbury and Scaifes Lagoon, Lake Wanaka, Otago, New Zealand. Rec Canterbury Mus 9: 309–336.
Chan YL, Anderson CN, Hadly EA . (2006). Bayesian estimation of the timing and severity of a population bottleneck from ancient DNA. PLoS Genet 2: e59.
Excoffier L, Dupanloup I, Huerta-Sanchez E, Sousa VC, Foll M . (2013). Robust demographic inference from genomic and SNP data. PLoS Genet 9: e1003905.
Fu Y-X, Chakraborty R . (1998). Simultaneous estimation of all the parameters of a stepwise mutation model. Genetics 150: 487–497.
Gamba C, Fernández E, Tirado M, Deguilloux M, Pemonge M, Utrilla P et al. (2012). Ancient DNA from an Early Neolithic Iberian population supports a pioneer colonization by first farmers. Mol Ecol 21: 45–56.
Ghirotto S, Mona S, Benazzo A, Paparazzo F, Caramelli D, Barbujani G . (2010). Inferring genealogical processes from patterns of bronze-age and modern DNA variation in Sardinia. Mol Biol Evol 27: 875–886.
Groombridge JJ, Jones CG, Bruford MW, Nichols RA . (2000). Conservation biology - 'Ghost' alleles of the Mauritius kestrel. Nature 403: 616–616.
Hardy OJ, Vekemans X . (2002). spagedi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2: 618–620.
Harper GL, Maclean N, Goulson D . (2006). Analysis of museum specimens suggests extreme genetic drift in the adonis blue butterfly (Polyommatus bellargus). Biol J Linnean Soc 88: 447–452.
Holdaway RN, Allentoft ME, Jacomb C, Oskam CL, Beavan NR, Bunce M . (2014). An extremely low-density human population exterminated New Zealand moa. Nat Commun 5: 5436.
Holdaway RN, Jacomb C . (2000). Rapid extinction of the moas (Aves: Dinorinthiformes): Model, test, and implications. Science 287: 2250–2254.
Huynen L, Gill BJ, Millar CD, Lambert DM . (2010). Ancient DNA reveals extreme egg morphology and nesting behavior in New Zealand's extinct moa. Proc Natl Acad Sci USA 107: 16201–16206.
Huynen L, Millar CD, Scofield RP, Lambert DM . (2003). Nuclear DNA sequences detect species limits in ancient moa. Nature 425: 175–178.
Ishida Y, Roca AL, Fratpietro S, Greenwood AD . (2012). Successful genotyping of microsatellites in the woolly mammoth. J Hered 103: 459–464.
Johnson JA, Bellinger MR, Toepfer JE, Dunn P . (2004). Temporal changes in allele frequencies and low effective population size in greater prairie-chickens. Mol Ecol 13: 2617–2630.
Kalinowski ST, Wagner AP, Taper ML . (2006). ML-RELATE: a computer program for maximum likelihood estimation of relatedness and relationship. Mol Ecol Notes 6: 576–579.
Kraemer P, Gerlach G . (2013). Demerelate: Functions to calculate relatedness on diploid genetic data. R package version 0.8-0 http://cran.rproject.org/package=Demerelate/index.html.
Miller CR, Joyce P, Waits LP . (2002). Assessing allelic dropout and genotype reliability using maximum likelihood. Genetics 160: 357–366.
Moore LA . (2007). Population ecology of the southern cassowary Casuarius casuarius johnsonii, Mission Beach north Queensland. J Ornithol 148: 357–366.
Navidi W, Arnheim N, Waterman MS . (1992). A multiple-tubes approach for accurate genotyping of very small DNA samples by using PCR - statistical considerations. Am J Human Genet 50: 347–359.
Nyström V, Humphrey J, Skoglund P, McKeown NJ, Vartanyan S, Shaw PW et al. (2012). Microsatellite genotyping reveals end-Pleistocene decline in mammoth autosomal genetic variation. Mol Ecol 21: 3391–3402.
Oliehoek PA, Windig JJ, van Arendonk JAM, Bijma P . (2006). Estimating relatedness between individuals in general populations with a focus on their use in conservation programs. Genetics 173: 483–496.
Petren K, Grant PR, Grant BR, Clack AA, Lescano NV . (2010). Multilocus genotypes from Charles Darwin's finches: biodiversity lost since the voyage of the Beagle. Philos Trans Royal Soc B-Biol Sci 365: 1009–1018.
Pritchard JK, Stephens M, Donnelly P . (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Queller DC, Goodnight KF . (1989). Estimating relatedness using genetic markers. Evolution 43: 258–275.
Queller DC, Strassmann JE, Hughes CR . (1993). Microsatellites and kinship. Trends Ecol Evol 8: 285–288.
Ramakrishnan U, Hadly EA . (2009). Using phylochronology to reveal cryptic population histories: review and synthesis of 29 ancient DNA studies. Mol Ecol 18: 1310–1330.
Roeder AD, Elsmore P, Greenhalgh M, McDonald A . (2009). Maximizing DNA profiling success from sub-optimal quantities of DNA: a staged approach. Forensic Sci Int Genet 3: 128–137.
Taberlet P, Griffin S, Goossens B, Questiau S, Manceau V, Escaravage N et al. (1996). Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res 24: 3189–3194.
Turvey ST, Holdaway RN . (2005). Postnatal ontogeny, population structure, and extinction of the giant moa Dinornis. J Morphol 265: 70–86.
White NE, Bunce M, Mawson PR, Dawson R, Saunders DA, Allentoft ME . (2014). Identifying conservation units after large‐scale land clearing: a spatio‐temporal molecular survey of endangered white‐tailed black cockatoos (Calyptorhynchus spp.). Divers Distrib 20: 1208–1220.
Whitehouse AM, Harley EH . (2001). Post-bottleneck genetic diversity of elephant populations in South Africa, revealed using microsatellite analysis. Mol Ecol 10: 2139–2149.
Wilson GA, Rannala B . (2003). Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163: 1177–1191.
Wood JR, Rawlence NJ, Rogers GM, Austin JJ, Worthy TH, Cooper A . (2008). Coprolite deposits reveal the diet and ecology of the extinct New Zealand megaherbivore moa (Aves, Dinornithiformes). Quat Sci Rev 27: 2593–2602.
Wood JR, Wilmshurst JM, Richardson SJ, Rawlence NJ, Wagstaff SJ, Worthy TH et al. (2013). Resolving lost herbivore community structure using coprolites of four sympatric moa species (Aves: Dinornithiformes). Proc Natl Acad Sci USA 110: 16910–16915.
Worthy TH, Holdaway RN . (2002) The lost world of the Moa. Canterbury University Press: Christchurch: Christchurch, New Zealand.
Acknowledgements
We are grateful to Malene Møhl and Charlotte Oskam for assistance with the sampling and to Marie Hale for advice on the analyses. We thank the Museum of New Zealand, Te Papa Tongarewa (A.J.D. Tennyson), Canterbury Museum (P. Scofield) and the American Museum of Natural History (C. Mehling). We thank Marcel Giesen and Sherwyn Veldhuizen (Bell Hill Vineyard), and the Hodgen (Pyramid Valley), and Earl (Rosslea) families for their support of our research. The research was primarily funded by Marsden Fund (Royal Society of New Zealand) contracts 06-PAL-001- and 09-UOO-164. M.E.A. is supported by the European Research Council, Marie Curie Actions, FP7 (Grant Agreement 300554). M.B. was supported in this research by an ARC future fellowship FT0991741.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
Allentoft, M., Heller, R., Holdaway, R. et al. Ancient DNA microsatellite analyses of the extinct New Zealand giant moa (Dinornis robustus) identify relatives within a single fossil site. Heredity 115, 481–487 (2015). https://doi.org/10.1038/hdy.2015.48
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2015.48