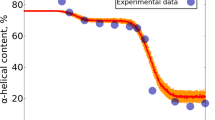
Abstract
The hydration of nonnative states is central to protein folding and stability but has been probed mainly by indirect methods. Here we use water 17O relaxation dispersion to monitor directly the internal and external hydration of α-lactalbumin, lysozyme, ribonuclease A, apomyoglobin and carbonic anhydrase in native and nonnative states. The results show that nonnative proteins are more structured and less solvent exposed than commonly believed. Molten globule proteins preserve most of the native internal hydration sites and have native-like surface hydration. Proteins denatured by guanidinium chloride are not fully solvent exposed but contain strongly perturbed occluded water. These findings shed new light on hydrophobic stabilization of proteins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Tanford, C. Protein denaturation. Adv. Protein Chem. 23, 121–282 (1968).
Kuwajima, K. The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins 6, 87–103 (1989).
Dill, K.A. & Shortle, D. Denatured states of proteins. Annu. Rev. Biochem. 60, 795–825 (1991).
Dobson, C.M. Unfolded proteins, compact states and molten globules. Curr. Opin. Struct. Biol. 2, 6–12 ( 1992).
Ptitsyn, O.B. in Protein folding (ed. Creighton, T.E.) 243–300 (W.H. Freeman & Co., New York; 1992).
Shortle, D. Denatured states of proteins and their roles in folding and stability. Curr. Opin. Struct. Biol. 3, 66–74 (1993).
Freire, E. Thermodynamics of partly folded intermediates in proteins. Annu. Rev. Biophys. Biomol. Struct. 24, 141– 165 (1995).
Fink, A.L. Compact intermediate states in protein folding. Annu. Rev. Biophys. Biomol. Struct. 24, 495–522 (1995).
Shortle, D. The denatured state (the other half of the folding equation) and its role in protein stability. FASEB J. 10, 27– 34 (1996).
Kuwajima, K. The molten globule state of α-lactalbumin. FASEB J. 10, 102–109 (1996).
Spolar, R.S., Livingstone, J.R. & Record, M.T. Use of liquid hydrocarbon and amide transfer data to estimate contributions to thermodynamic functions of protein folding from the removal of nonpolar and polar surface from water. Biochemistry 31, 3947–3955 ( 1992).
Makhatadze, G.I. & Privalov, P.L. Hydration effects in protein unfolding. Biophys. Chem. 51, 291–309 (1994).
Gast, K., Zirwer, D., Welfle, H., Bychkova, V.E. & Ptitsyn, O.B. Quasielastic light scattering from human α-lactalbumin: comparison of molecular dimensions in native and molten globule states. Int. J. Biol. Macromol. 8, 231–236 (1986).
Kataoka, M., Kuwajima, K., Tokunaga, F. & Goto, Y. Structural characterization of the molten globule of α-lactalbumin by solution X-ray scattering. Protein Sci. 6, 422– 430 (1997).
Gast, K. et al. Compactness of protein molten globules: temperature-induced structural changes of the apomyoglobin folding intermediate. Eur. Biophys. J. 23, 297–305 ( 1994).
Kataoka, M. et al. Structural characterization of the molten globule and native states of apomyoglobin by solution X-ray scattering. J. Mol. Biol. 249, 215–228 ( 1995).
Nishii, I., Kataoka, M., Tokunaga, F. & Goto, Y. Cold denaturation of the molten globule states of apomyoglobin and a profile for protein folding. Biochemistry 33, 4903– 4909 (1994).
Uchiyama, H., Perez-Prat, E.M., Watanabe, K., Kumagai, I. & Kuwajima, K. Effects of amino acid substitutions in the hydrophobic core of α-lactalbumin on the stability of the molten globule state. Protein Eng. 8, 1153– 1161 (1995).
Kharakoz, D.P. & Bychkova, V.E. Molten globule of human α-lactalbumin: hydration, density, and compressibility of the interior. Biochemistry 36, 1882– 1890 (1997).
Finkelstein, A.V. & Shakhnovich, E.I. Theory of cooperative transitions in protein molecules. II. Phase diagram for a protein molecule in solution. Biopolymers 28, 1681 –1694 (1989).
Alonso, D.O.V., Dill, K.A. & Stigter, D. The three states of globular proteins: acid denaturation. Biopolymers 31, 1631–1649 (1991).
Williams, M.A., Thornton, J.M. & Goodfellow, J.M. Modelling protein unfolding: hen egg-white lysozyme. Protein Eng. 10, 895–903 (1997).
Kunz, I.D. & Kauzmann, W. Hydration of proteins and polypeptides. Adv. Protein Chem. 28, 239– 345 (1974).
Otting, G. NMR studies of water bound to biological molecules. Prog. NMR Spectrosc. 31, 259–285 ( 1997).
Halle, B., Denisov, V.P. & Venu, K. in Modern techniques in protein NMR Vol. 17 (eds Berliner, L.J. & Krishna, N.R.) in the press (Plenum Press, New York; 1999).
Denisov, V.P. & Halle, B. Thermal denaturation of ribonuclease A characterized by water 17O and 2H magnetic relaxation dispersion. Biochemistry 37, 9595–9604 (1998).
Denisov, V.P. & Halle, B. Protein hydration dynamics in aqueous solution. Faraday Discuss. 103, 227– 244 (1996).
Denisov, V.P., Peters, J., Hörlein, H.D. & Halle, B. Using buried water molecules to explore the energy landscape of proteins. Nature Struct. Biol. 3, 505– 509 (1996).
Denisov, V.P. & Halle, B. Direct observation of calcium-coordinated water in calbindin D9k by nuclear magnetic relaxation dispersion. J. Am. Chem. Soc. 117, 8456– 8465 (1995).
Ishimura, M. & Uedaira, H. Natural-abundance oxygen-17 magnetic relaxation in aqueous solutions of apolar amino acids and glycine peptides. Bull Chem. Soc. Jpn. 63, 1– 5 (1990).
Gerstein, M. & Chothia, C. Packing at the protein-water interface. Proc. Natl. Acad. Sci. USA 93, 10167– 10172 (1996).
Edelhoch, H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 6, 1948–1954 (1967).
Pace, C.N. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 131, 266– 280 (1986).
Kuwajima, K., Nitta, K., Yoneyama, M. & Sugai, S. Three-state denaturation of α-lactalbumin by guanidine hydrochloride. J. Mol. Biol. 106, 359–373 ( 1976).
Schellman, J.A. The thermodynamics of solvent exchange. Biopolymers 34, 1015–1026 (1994).
Schellman, J.A. A simple model for solvation in mixed solvents. Application to the stabilization and destabilization of macromolecular structures. Biophys. Chem. 37, 121–140 ( 1990).
Makhatadze, G.I. & Privalov, P.L. Protein interactions with urea and guanidinium chloride. A calorimetric study. J. Mol. Biol. 226, 491–505 ( 1992).
Schellman, J.A. & Gassner, N.C. The enthalpy of transfer of unfolded proteins into solutions of urea and guanidinium chloride. Biophys. Chem. 59, 259– 275 (1996).
Schulman, B.A., Kim, P.S., Dobson, C.M. & Redfield, C. A residue-specific NMR view of the non-cooperative unfolding of a molten globule. Nature Struct. Biol. 4, 630–634 (1997).
Dolgikh, D.A. et al. Compact state of a protein molecule with pronounced small-scale mobility: bovine α-lactalbumin. Eur. Biophys. J. 13, 109–121 (1985).
Kronman, M.J. Metal-ion binding and the molecular conformational properties of α-lactalbumin. Crit. Rev. Biochem. Mol. Biol. 24, 565– 667 (1989).
Wu, L.C., Peng, Z. & Kim, P.S. Bipartite structure of the α-lactalbumin molten globule. Nature Struct. Biol. 2, 281–286 (1995).
Lecomte, J.T.J., Kao, Y.H. & Cocco, M.J. The native state of apomyoglobin described by proton NMR spectroscopy: the A-B-G-H interface of wild-type sperm whale apomyoglobin. Proteins 25, 267–285 (1996).
Eliezer, D., Yao, J., Dyson, H.J. & Wright, P.E. Structural and dynamic characterization of partially folded states of apomyoglobin and implications for protein folding. Nature Struct. Biol. 5, 148–155 (1998).
Mårtenson, L.-G. et al. Characterization of folding intermediates of human carbonic anhydrase II: Probing substructure by chemical labeling of SH groups introduced by site-directed mutagenesis. Biochemistry 32, 224–231 (1993).
Svensson, M. et al. Mapping the folding intermediate of human carbonic anhydrase II. Probing substructure by chemical reactivity and spin and fluorescence labeling of engineered cysteine residues. Biochemistry 34, 8606–8620 (1995).
Halle, B., Jóhannesson, H. & Venu, K. Model-free analysis of stretched relaxation dispersions. J. Magn. Reson. 135, 1– 13 (1998).
Barron, L.D., Hecht, L. & Wilson, G. The lubricant of life: a proposal that solvent water promotes extremely fast conformational fluctuations in mobile heteropolypeptide structure. Biochemistry 36, 13143– 13147 (1997).
Damaschun, G., Gernat, C., Damaschun, H., Bychkova, V.E. & Ptitsyn, O.B. Comparison of intramolecular packing of a protein in native and 'molten globule' states. Int. J. Biol. Macromol. 8, 226–230 ( 1986).
Pfeil, W. Is the molten globule a third thermodynamic state of protein? The example of α-lactalbumin. Proteins 30, 43– 48 (1998).
Griko, Y.V. & Privalov, P.L. Thermodynamic puzzle of apomyoglobin unfolding. J. Mol. Biol. 235, 1318– 1325 (1994).
Hinz, H.-J., Vogl, T. & Meyer, R. An alternative interpretation of the heat capacity changes associated with protein unfolding. Biophys. Chem. 52, 275 –285 (1994).
Yang, D., Mok, Y.-K., Forman-Kay, J.D., Farrow, N.A. & Kay, L.E. Contributions to protein entropy and heat capacity from bond vector motions measured by NMR spin relaxation. J. Mol. Biol. 272, 790–804 (1997).
Matthews, C.R. Pathways of protein folding. Annu. Rev. Biochem. 62 , 653–683 (1993).
Ptitsyn, O.B. Structures of folding intermediates. Curr. Opin. Struct. Biol. 5, 74–78 (1995 ).
Shortle, D., Wang, Y., Gillespie, J.R. & Wrabl, J.O. Protein folding for realists: a timeless phenomenon. Protein Sci. 5, 991–1000 ( 1996).
Miranker, A.D. & Dobson, C.M. Collapse and cooperativity in protein folding. Curr. Opin. Struct. Biol. 6, 31–42 (1996).
Aronsson, G., Mårtensson, L.-G., Carlsson, U. & Jonsson, B.-H. Folding and stability of the N-terminus of human carbonic anhydrase II. Biochemistry 34, 2153–2162 (1995).
Teale, F.W.J. Cleavage of the haem–protein link by acid methylethylketone. Biochim. Biophys. Acta 35, 543 (1959).
Hiraoka, Y. & Sugai, S. Equilibrium and kinetic study of sodium- and potassium-induced conformational changes of apo-α-lactalbumin. Int. J. Pept. Protein Res. 26, 252– 261 (1985).
Hughson, F.M., Wright, P.E. & Baldwin, R.L. Structural characterization of a partly folded apomyoglobin intermediate. Science 249, 1544– 1548 (1990).
Wong, K.P. & Hamlin, L.M. Acid denaturation of bovine carbonic anhydrase B. Biochemistry 13, 2678– 2683 (1974).
Jagannadham, M.V. & Balasubramanian, D. The molten globular intermediate form in the folding pathway of human carbonic anhydrase B. FEBS Lett. 188, 326– 330 (1985).
Peng, Z. & Kim, P.S. A protein dissection study of a molten globule. Biochemistry 33, 2136– 2141 (1994).
Loh, S.N., Kay, M.S. & Baldwin, R.L. Structure and stability of a second molten globule intermediate in the apomyoglobin folding pathway. Proc. Natl. Acad. Sci. USA 92, 5446–5450 ( 1995).
Denisov, V.P. & Halle, B. Protein hydration dynamics in aqueous solution. A comparison of bovine pancreatic trypsin inhibitor and ubiquitin by oxygen-17 spin relaxation dispersion. J. Mol. Biol. 245, 682–697 (1995).
Myers, J.K., Pace, C.N. & Scholtz, J.M. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 4, 2138–2148 (1995).
Acknowledgements
We thank E. Thulin for help with protein purification. This work was supported by the Swedish Natural Science Research Council (NFR) and the Royal Swedish Academy of Sciences (KVA).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Denisov, V., Jonsson, BH. & Halle, B. Hydration of denatured and molten globule proteins. Nat Struct Mol Biol 6, 253–260 (1999). https://doi.org/10.1038/6692
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/6692
This article is cited by
-
High protein flexibility and reduced hydration water dynamics are key pressure adaptive strategies in prokaryotes
Scientific Reports (2016)
-
Insight of Endo-1,4-Xylanase II from Trichoderma reesei: Conserved Water-Mediated H-Bond and Ion Pairs Interactions
The Protein Journal (2013)
-
How to tailor heat-induced whey protein/κ-casein complexes as a means to investigate the acid gelation of milk—a review
Dairy Science & Technology (2011)
-
Hydration Profiles of Amyloidogenic Molecular Structures
Journal of Biological Physics (2008)
-
The Partial Molar Volume and Heat Capacity of the Glycyl Group in Aqueous Solution at 25^C
Journal of Solution Chemistry (2006)