Abstract
MicroRNAs (miRNA) that guide sequence-specific posttranscriptional gene silencing play an important role in gene expression required for both developmental processes and responses to environmental conditions in plants. However, little is known about the transcriptional and posttranscriptional regulation of miRNA expression. Histone acetylation plays an important role in chromatin remodeling and is required for gene activation. By analyzing the accumulation of subset of miRNAs and the corresponding primary miRNAs in mutants of Arabidopsis, we show that histone acetyltransferase GCN5 (General control non-repressed protein5) has a general repressive effect on miRNA production, while it is required for the expression of a subset of (e.g. stress-inducible) MIRNA genes. The general negative function of GCN5 in miRNA production is likely achieved through an indirect repression of the miRNA machinery genes such as DICER LIKE1 (DCL1), SERRATE (SE), HYPONASTIC LEAVES1 (HYL1) and ARGONAUTE1 (AGO1). Chromatin immunoprecipitation assays revealed that GCN5 targets to a subset of MIRNA genes and is required for acetylation of histone H3 lysine 14 at these loci. Moreover, inhibition of histone deacetylation by trichostatin A treatment or in histone deacetylase gene mutants impaired the accumulation of certain miRNAs. These data together suggest that Arabidopsis GCN5 interferes with the miRNA pathway at both the transcriptional and posttranscriptional levels and histone acetylation/deacetylation is an epigenetic mechanism involved in the regulation of miRNA production.
Similar content being viewed by others
Introduction
Histone modification plays an important role in chromatin remodeling and gene expression. Among the modifications, acetylation/deacetylation appears as a key switch for inter-conversion between permissive and repressive states of chromatin domains important for regulating genome activity such as gene transcription. Hyper-acetylation of histones relaxes chromatin structure and is usually associated with transcriptional activation, whereas hypo-acetylation induces chromatin compaction and gene repression 1. The homeostatic balance of histone acetylation in chromatin is maintained by antagonistic actions of histone acetyltransferases (HAT) and histone deacetylases (HDAC). The identification of the yeast (Saccharomyces cerevisiae) transcriptional regulator GCN5 (General control non-repressed protein5) as a HAT suggests that histone acetylation is tightly linked to gene transcriptional regulation 2. GCN5 plays an essential role in gene expression, as deletion mutations of GCN5 result in either up- or downregulation of a large number of genes in yeast 3. Drosophila also appears to require GCN5 for normal developmental processes, as flies harboring a null allele display defects in metamorphosis and oogenesis 4. Knockouts of mouse GCN5 result in lethality approximately one third of the way through embryonic development 5, 6.
GCN5 is evolutionarily conserved. An ortholog found in Arabidopsis is also called HAG1 7. T-DNA insertion mutations of Arabidopsis GCN5 induce pleiotropic defects that alter many aspects of plant development and responses to environmental conditions, such as light and cold 8, 9, 10. Arabidopsis GCN5 found to be involved mostly in the acetylation of histone H3K9, K14 and K27, is shown to be required for the expression of a large number of genes 8, 11, 12, suggesting that this protein is involved in both long-term epigenetic regulation of chromatin modification and short-term control of transcriptional switches. GCN5 appears to be a major HAT required for gene expression in Arabidopsis, as mutations of other members of the Arabidopsis HAT family produce relatively moderate effects 13.
Gene expression is also controlled by microRNAs (miRNAs) that guide sequence-specific posttranscriptional gene silencing. miRNAs are encoded by specific genes in the genome, which are transcribed as primary transcripts called primary miRNAs (pri-miRNAs). In Arabidopsis, pri-miRNAs are processed to mature miRNAs by the enzyme DICER-LIKE1 (DCL1) and other proteins including the zinc-finger domain protein SERRATE (SE) and a double-stranded RNA-binding protein, HYPONASTIC LEAVES1 (HYL1) 14, 15, 16. In addition, plant miRNAs are methylated by HUA ENHANCER1 (HEN1) at the 3′-ends and are loaded onto ARGONAUTE1 (AGO1) to target the homologous mRNA 17, 18. miRNAs are involved in the control of various plant developmental processes, such as leaf morphogenesis 19, floral development 20, root development 21 and developmental phase transitions 22, etc. Many miRNAs regulate plant development by delimiting the regions of accumulation of transcripts encoding transcription factors that function in development. Mutants defective in the accumulation of miRNAs, such as dcl1, hyl1, hen, se and ago1, produce pleiotropic developmental phenotypes 14, 15, 16, 17, 18. Importantly, plant miRNAs also mediate stress responses. Several recent studies show that miRNAs play important roles in responding to environmental conditions 23, 24, 25, 26, 27, 28, and are inducible by hormones or growth conditions. For instance, miR159 levels are enhanced by gibberellins 29, miR164 is transiently induced by auxin treatments 21; miR395 is highly induced by sulfate starvation and miR399 is strongly and specifically induced by low-phosphate conditions 23, 30. Therefore, the regulation of miRNA production is an important regulatory aspect of plant gene expression.
Overall, histone acetylation and the miRNA are opposite regulatory pathways of gene expression. However, little is known about the interaction between the two pathways. Here, we show that GCN5-dependent histone acetylation interferes with the miRNA pathways by regulating miRNA production at several levels.
Results
Analysis of miRNA accumulations in gcn5 mutants
To examine whether GCN5 is involved in the regulation of miRNA accumulation, total RNAs isolated from inflorescences of wild-type and the previously characterized T-DNA insertion mutant gcn5-2 10 were analyzed by Northern blots using 32P-labled oligonucleotides complementary to 11 Arabidopsis miRNAs (miR) as probes (Supplementary information, Table S1). The target genes of these miRNAs are involved in plant development, miRNA metabolism itself and responses to environmental conditions (Table 1). The same blots were hybridized with a probe to reveal the heterogeneous nuclear RNA U6 as loading controls. The level of most of the tested miRNAs was increased in gcn5-2 compared to wild-type plants (Figure 1). The increase of some of the miRNAs (e.g. miR159, miR172 and miR399) was more pronounced than others (e.g. miR165). In contrast, the level of miR164 was actually decreased in the mutant, while miR395 was not detected in either wild type or the mutant (not shown), possibly because it is expressed only under low-sulfate conditions 30.
Analysis of miRNAs accumulation in gcn5. Total RNA isolated from the wild type (WT) and gcn5-2 inflorescences were analyzed by northern hybridizations using probes of ten representative miRNAs as indicated on the left of the panels. The same blots were hybridized with the probe of U6 snRNA. Relative accumulations of miRNAs in gcn5-2 compared to wild type are indicated.
Analysis of pri-miRNA levels in gcn5 mutants
In order to know whether the increased accumulation of miRNAs in the gcn5 mutant was due to increased expression of the corresponding MIRNA genes or an enhanced maturation process from the precursors, levels of the corresponding pri-miRNAs were measured by reverse transcription-polymerase chain reaction (RT-PCR) using gene-specific primers. The miRNAs under study are encoded by multiple genes. One member (that is previously shown to be expressed, see Table 1) per gene family was selected for the analysis. Both gel-based and real-time PCR analyses were performed. The results are presented in Figure 2 and Supplementary information, Figure S1. The level of 7 tested pri-miRNAs (pri-miR156a, 164a, 165a, 168a, 172a, 395e and 399d) was decreased in gcn5-2, and that of the remaining 3 pri-miRNAs (pri-miR157c, 159a and 162a) was not reduced. Analysis of other members of the gene families (i.e., pri-miR165e, 157a and 164c) revealed similar results (Supplementary information, Figure S1). In addition, the decrease of pri-miR172a, 395e and 399d was observed in both seedlings and inflorescences of gcn5-2 (Supplementary information, Figure S2). However, not all of the gene members of pri-miR172, 395 and 399 families were affected by the gcn5 mutation (Supplementary information, Figure S2). The increase of most of the tested miRNAs coincided with the decrease of the corresponding pri-miRNAs, suggesting that the maturation process of these miRNAs might be enhanced in the mutant, while a possible downregulation of these MIRNA genes in the mutant is not excluded. Unchanged or increased levels of pri-miR157a, pri-miR159a and pri-miR162a suggest that either the expression of the MIRNA genes was increased in the mutant or the increased miRNA levels were originated from other pri-miRNA gene members. As miR164 was reduced in the mutants, the decrease of pri-miR164a suggests that the expression of the gene was repressed in the mutant. Although miR395 was not detected in either wild-type or gcn5 mutants, the pri-miR395e was found to be reduced in the mutant (Figure 2; Supplementary information, Figure S2). Together, the data suggest that GCN5 may be involved in the regulation of both pri-miRNA maturation and MIRNA gene expression.
Analysis of pri-miRNA levels in gcn5. Gene-specific pri-miRNAs were detected by real-time RT-PCR analysis of wild-type and gcn5-2 inflorescence total RNAs. The pri-miRNA levels (relative to that of EF1 transcripts) in wild type were set as 100. Error bars represent SD from at least four repetitions.
Expression of miRNA targets in gcn5 mutants
RT-PCR analysis was performed to study whether the altered miRNAs affected the accumulation of the target mRNAs in the gcn5-1 and gcn5-2 backgrounds. Gene-specific primers corresponding to targets of the miRNA were used for the analysis. Compared with the wild type, the accumulation of five target mRNAs (i.e., DCL1; AGO1; CUP-SHAPED COTYLEDON1 (CUC1); MYB33 and PHOSPHATE HOMEOSTASIS2 (PHO2)) was increased, four (i.e., Squamosa promoter-binding protein (SBP)-LIKE PROTEIN10 (SLP10), AUXIN-RESPONSIVE FACTOR6 (ARF6), APETALA2 (AP2) and PHABULOSA (PHB)) unchanged and one (i.e., SPL3) decreased in the mutants (Figure 3A). The increase of CUC1 mRNA accumulation in the mutants was in agreement with the decrease of miR164 in gcn5-2. The decrease of SPL3 mRNA could be explained by the accumulation of miR157/156 (which target both SPL3 and SPL10). However, the level of the remaining targets, albeit with elevated levels of the corresponding miRNAs, was either increased or unchanged in the mutants (Figure 3A). This observation may suggest that either the increased miRNAs did not lead to additional degradation of the target mRNAs or the decreased mRNA levels were compensated by increased transcription of the genes as a consequence of the gcn5 mutation. In addition, although miR395 was not detected in the wild-type or the mutant plants (not shown), the level of its target mRNA (ATP Sulphurylase4, APS4) was decreased (Figure 3A). These observations suggest that transcription of many of the miRNA target genes might also be regulated by GCN5.
Target gene expression of miRNAs in wild-type and gcn5 plants. (A) RT-PCR detection of target transcripts of ten miRNAs (see Table 1) in wild-type (WT) and in gcn5-1- and gcn5-2-mutant inflorescence total RNAs. PCR reactions without reverse transcriptase (RT) were included as controls. PCR cycle numbers are indicated on the right. Actin transcripts were detected as quantity controls. (B) RT-PCR detection of SE, HYL1 and HEN1 mRNA levels. (C) Detection of SE, HYL1 and HEN1 mRNA levels by Northern hybridizations. 18S rRNA was revealed as loading control. Relation accumulations of the transcripts in the mutants compared to wild type are indicated.
Interestingly, DCL1 and AGO1 mRNAs were among the miRNA targets with increased levels in the gcn5 mutant. Since these two genes encode components of the miRNA machinery, we tested whether the other components of the machinery were also deregulated in the mutants. RT-PCR and Northern blot analysis showed that the transcripts of SE and HYL were also increased in the mutants (Figure 3B and 3C). HEN1 mRNA level was not clearly changed. These data suggest that GCN5 may have a repressive function to negatively regulate the expression of several components of the miRNA machinery.
Tests of cleavage of miRNA targets
In order to evaluate the impact of altered miRNA levels on the accumulation of target mRNAs, we tested the cleaved products of 5 transcripts (i.e., DCL1, AGO1, MYB33, AP2 and PHB) by detecting the exposed 5′ RNA ends according to Llave et al. 31. For MYB33, AGO1 and DCL1, cleavage products were clearly more abundant in gcn5-2 than in wild type, suggesting that the accumulation of miR159, miR162 and miR168 in gcn5-2 resulted in more cleavage of the targets than in wild type (Figure 4). The abundance of cleavage products of PHB and AP2 mRNAs was slightly higher in the mutant than in the wild type. This could be explained by only a moderate increase of miR165 and by the fact that miR172 functions by inhibiting translation of the target mRNA 20.
gcn5 mutation increases the accumulation of miRNA cleavage products. The nested 5′ race RT-PCR products of 5 miRNA cleavage targets (i.e., DCL1, AGO1, MYB33, AP2 and PHB). Ubiquitin (UBQ) and actin (ACT) transcripts were detected by RT-PCR as quantity controls. PCR reactions without RNA (H2O) were included as negative controls. PCR cycle numbers are indicated on the right of the panels.
Some of the miRNA genes are direct targets of GCN5
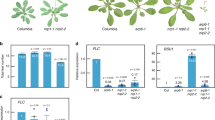
The above data suggest that GCN5 is involved in the expression of components of the miRNA pathway and of some MIRNA genes. As GCN5 is supposed to have an activating function for gene transcription, the negative function of GCN5 in the expression of components of the miRNA machinery suggests that GCN5 may not directly regulate these genes. Chromatin immunoprecipitation (ChIP) assays with antibodies against Arabidopsis GCN5 protein suggested that GCN5 was not directly bound to these genes (Supplementary information, Figure S3). A similar analysis of 10 MIRNA loci revealed that GCN5 was associated with the genes of pri-miR165a, 172a, 395e and 399d in the wild-type plants (Figure 5). In gcn5-2, the association was much reduced (Figure 5A), suggesting that these MIRNA genes may be direct targets of GCN5. As GCN5 is required for acetylation of histone H3 lysine 14 (H3K14) 9, 32, ChIP assays were performed with antibodies against acetylated H3K14. The analysis revealed that in the mutants H3K14 acetylation was reduced mainly on the GCN5 targets (Figure 5B).
GCN5 directly targets a subset of MIRNA genes and is required for acetylation of histone H3 lysine 14. (A) GCN5 is associated with a subset of MIRNA loci. Chromatin fragments isolated from wild-type (WT) and gcn5 seedling nuclei were immunoprecipitated by antibodies against GCN5. Both the input chromatin and the precipitated products were analyzed by real-time PCR using the primer sets corresponding to 10 MIRNA loci (Supplementary information, Table S2). The percentages of precipitated chromatin products from both wild type and the mutant are shown (Y axis). Error bars represent SD from at least three repetitions. (B) Histone H3 lysine 14 (H3K14) acetylation of a subset of MIRNA loci is reduced in gcn5 plants. Chromatin fragments isolated from wild-type (WT) and gcn5 seedling nuclei were immunoprecipitated by antibodies against acetylated H3K14. Both the input chromatin and the precipitated products were analyzed by real-time PCR using the same primer sets as in (A). The percentages of precipitated chromatin products from both wild type and the mutant are shown (Y axis). Error bars represent SD from at least three repetitions.
Analysis of gcn5 serrate double mutants
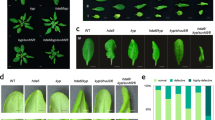
The serrate (se) mutation produces a highly pleiotropic phenotype including leaf margin development (serration), phyllotaxy arrangement of lateral structures such as leaf and flowers and random internode lengths between adjacent flowers, in addition to altered floral organ numbers 33 (Figure 6A). The gcn5 mutants also show a pleiotropic phenotype including upward curled leaves, apical dominance and altered floral organ numbers 10 (Figure 6A). SE mRNA was accumulated in gcn5 mutants (Figure 3B and 3C). We wondered whether the se phenotype could be attenuated by the gcn5 mutation. However, the gcn5 se double mutants displayed a severer phenotype than the single mutants (Figure 6A). First, the characteristic leaf serration phenotype of se was enhanced in the double mutants that produced highly irregular leaf margins (Figure 6A). It has been shown that the balance between miR164 and CUC1/2 genes controls leaf margin serration in Arabidopsis 34. The severer leaf margin phenotype of the double mutants might be explained by the reduced accumulation of miR164 in both gcn5 and se. Other phenotypes of the double mutants include reduced distance between adjacent flowers, stem fasciation, disorganized floral shape with highly variable floral organ numbers and malformed floral organs (Figure 6A). The double mutants were totally infertile. The severer phenotype of the double mutants might be a consequence of loss-of-function of the two genes involved in such two important gene regulatory pathways (i.e., histone acetylation and miRNA). As the expression of SE is not constitutive, but restricted to specific tissue types and developmental stages, and ectopic expression of SE induces a variety of phenotypes including variable floral organ numbers and altered floral organ morphology 33, the induction of SE expression in gcn5 might be ectopic rather than simply restore the native SE expression pattern.
Analysis of serrate (se) and gcn5-2 double mutations. (A) Phenotype comparison of plants with se and gcn5 single and double mutations. Leaf (upper panels), flower (middle panels) and inflorescence (lower panels) of se, se gcn5-2 and gcn5-2 were compared to wild type (WT). (B) Comparison of miRNA levels in serrate with that in the double mutants. miRNA were detected from total RNAs isolated from inflorescences by Northern hybridizations using the probes indicated on the left of the panels.
Examination of miRNA accumulation revealed that the double mutation increased the levels of miR159, miR162, miR167 and miR168 compared to the single se mutation. In agreement with the repression of miR164 accumulation by the single mutations, miR164 was not increased in the double mutants. Therefore, compared with se single mutation, the gcn5 se double mutations restored the accumulation of a subset of miRNAs.
Effect of inhibition of histone deacetylation on miRNA accumulation
To evaluate if histone acetylation was a general mechanism involved in miRNA accumulation, seedlings were treated with trichostatin A (TSA), a HDAC inhibitor, and analyzed by Northern blots using the miR157 probe. At 6 h after the treatment, the level of miR157 was decreased compared to the control (Figure 7). To further study the role of HDACs in miRNA accumulation, two T-DNA insertion mutants of HDAC genes were used for analysis. The mutation of hda9 did not produce any morphological phenotype (not shown), while hd1 (or hda19) showed pleiotropic defects 35. Both HDA9 and HDA19 are members of the RPD3 (REDUCED POTASSIUM DEPENDANT3)-type of HDAC genes 7. As shown in Figure 7B, the accumulation of 3 tested miRNAs (i.e., miR157, miR162 and miR172) was reduced in hda9 mutants. However, only miR157 showed some reduction in hd1.
Effects of histone deacetylation inhibition on miRNA accumulation. (A) Northern blot detection of miR157 levels in plants treated by trichostatin A (TSA) or DMSO (mock) during 0 or 6 h. U6 snRNA was detected as controls. Quantifications of the miRNA signals relative to U6 RNA (with that at 0 h assessed as 100) are indicated on the right. (B) Northern blot detection of 4 miRNAs in mutant plants of histone deacetylase genes HDA9 and HD1 (HDA19) in comparison with wild type (WT). Relative signals (with that in wild type assessed as 100) are indicated below the gels.
Discussion
GCN5 is involved in the regulation of miRNA accumulation at different levels
Transcription, processing and loading to AGO-containing RNA-induced silencing complexes determine mature miRNA levels found in cells. Little is known about the transcriptional and posttranscriptional regulation of miRNA expression, although promoter activity of a few miRNA genes has been described 36, 37. Here, we provide evidence that Arabidopsis GCN5 has a general repressive effect on miRNA production. Because in most cases, the increase of miRNA levels in gcn5 mutants was associated with a decrease of the corresponding pri-miRNAs, GCN5 is likely to interfere with the miRNA pathway at the level of miRNA processing. This hypothesis is supported by increased expression of genes encoding key components of the miRNA machinery (i.e., DCL1, SE, HYL and AGO1) in gcn5 mutants (Figure 3). The co-regulation of these key components suggests the possibility that their genes may share common regulatory elements responding to GCN5 activity. However, the repressive effect of GCN5 on miRNA machinery genes is likely to be indirect, as no direct binding of GCN5 to the promoters was detected in this work and in a more large-scale study (Supplementary information, Figure S1) 12. We speculate that there may exist a common repressor of the miRNA machinery genes, the activity of which is dependent on GCN5. At this stage, it is not, however, excluded that GCN5 may also mediate the repression of miRNA processing through a different mechanism, as the accumulation of tested miRNAs was differentially affected by the gcn5 mutation (Figure 1). In addition, the restoration of levels of most of the tested miRNAs in gcn5 se double mutants shown in Figure 6B suggests that the repressive effect of GCN5 on miRNA accumulation is not fully dependent on the repression of SE. GCN5 is an activator of transcription of a large number of genes. The present data suggest that GCN5 has an additional means to promote gene expression through inhibition of miRNA accumulation.
The present data suggest that GCN5 is also required for the expression of a subset of MIRNA genes. For instance, the concurrent decrease of both the mature miR164 and the corresponding pri-miRNA indicates that GCN5 is required for the MIRNA gene transcription. However, GCN5 is likely to regulate this MIRNA indirectly, as no direct association of GCN5 with the gene was detected (Figure 5A). In addition, the differential accumulation of pri-miRNAs found in gcn5 mutants might be due to altered transcription rate of the MIRNA genes in the mutants, although this could also be caused by differential maturation processes of the pri-miRNAs.
Importantly, we have shown that GCN5 is associated with 4 of the 10 tested MIRNA loci, indicating that MIRNA genes are amongst the genome-wide targets of GCN5. It is recently shown that GCN5 associated with 40% of the tested gene promoters 12. At most sites, GCN5 binding does not depend on the integrity of its bromodomain that is required for binding to only 11% of the target promoters and for acetylation of H3K14 on these promoters 12. Interestingly, the binding to MIRNA genes seemed to require the bromodomain of GCN5, as the gcn5-2 mutation, which deletes the C-terminal bromodomain of the protein 10, 12, clearly reduced the association of GCN5 and H3K14 acetylation on the loci (Figure 5). The expression of these genes (i.e., pri-miR levels) was clearly reduced in the gcn5-2 mutant (Figure 2), demonstrating a correlation between H3K14 acetylation and the MIRNA gene expression. These observations suggest that GCN5-dependent H3K14 acetylation is required for the expression of a subset of MIRNA genes.
We noticed that miRNAs encoded by the four GCN5 targets have been shown to be responsive to environmental conditions. miR165 and miR172 are inducible by cold stress 26, whereas miR395 and miR399 are highly induced under sulfate and phosphate starvation, respectively 27, 28, 30. Although it is not known whether the stress-induction of miRNA accumulation is at the transcriptional or posttranscriptional level, the present data suggest that the GCN5 may have a function in stress-induction of MIRNA genes expression. As GCN5 has been shown to be involved in plant response to environmental conditions 8, 12, 32, its involvement in stress-responsive miRNA accumulation highlights the importance of GCN5 for plant adaptation to environmental conditions.
Taken together, the present data show that Arabidopsis GCN5 regulates miRNA accumulation at both transcriptional and posttranscriptional levels.
Histone acetylation is an epigenetic mechanism regulating miRNA accumulation
Epigenetic silencing of miRNA expression has been reported in mammalian cells, but the underlying mechanism is unknown 38. Histone modification is thought to be one of the mechanisms of epigenetic regulation of gene expression. Our data suggest that histone acetylation is an important factor to regulate miRNA accumulation. We have shown that treatment with the general HDAC inhibitor TSA reduced the accumulation of a miRNA (miR157) (Figure 7). A study of breast cancer cells treated with another HDAC inhibitor (i.e., Hydroxamic acid) shows that about 40% of the more than 60 examined miRNA species were downregulated by the treatment 39. In addition, accumulation of a subset of miRNAs was decreased in hda19/hd1 and hda9 mutants (Figure 7). HDA19/AtHD1 has been shown to be a general HDAC required for the expression of a large number of genes and functions together with GCN5 as an antagonistic couple in a few regulatory pathways 9, 35. However, HDA19 seemed to play a less important role than HDA9 in MIRNA gene expression (Figure 7), indicating that HDAC genes may have different functions in MIRNA gene regulation. Taken together, our data suggest that histone acetylation/deacteylation may be an important epigenetic mechanism for the regulation of miRNA accumulation in Arabidopsis.
Materials and Methods
Plant material
Arabidopsis thaliana ecotype Wassilewskija (Ws, gcn5-1 and gcn5-2) was grown under 16-h light and 8-h dark condition at 21 °C for 12 days and transferred on soil. For trichostatin A treatment, plants were grown on 1/2MS media supplemented with 5 μM TSA (Sigma-Aldrich) dissolved in dimethyl sulfoxide (DMSO) or supplemented with DMSO only.
Northern blotting for miRNA
RNA isolation from inflorescence and seedlings was performed using TRIzol® reagent (Invitrogen). A total of 10 μg of RNA was used for migration on 17% denaturing polyacrylamide gels. Oligonucleotides used as probes are listed in Table S1. Probes for miRNA were end-labeled with γ32P-ATP using polynucleotide kinase. The same blots were also analyzed with an oligonucleotide probe specific to snRNA U6 as a loading control.
Detection of pri-miRNA and expression of target genes by RT-PCR
Total RNA was isolated from inflorescence tissues of wild-type and gcn5 mutants using Nucleospin® RNA plant kit (Macherey-Nagel). cDNA was synthesized from 1 μg of RNA by ImProm-II™ Reverse Transcriptase (Promega) and analyzed by both real-time and gel-based PCR. For real-time PCR, the reactions were performed in a 96-well plate with a Light Cycler 480 system (Roche), using SYBR Green to monitor dsDNA synthesis. Reactions contained 10 μl 2× SYBR Green Master Mix reagent (Roche), 1 μl of cDNA (1 ng/μl) and 200 nM of each gene-specific primer in a final volume of 20 μl. The following standard thermal profile was used for all PCR reactions: 50 °C for 2 min, 95 °C for 10 min, 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Amplicon dissociation curves, i.e., melting curves, were recorded after 40 cycles by heating from 60 °C to 95 °C with a ramp speed of 1.9 °C/min. Data were analyzed using the Light Cycler 480 software 1.5.0 (Roche). Primers for detecting pri-miR157c, pri-miR159a, pri-miR164a have been described by Grigg et al. 40. EF1 transcripts were measured as internal reference controls as described earlier 41. Primers for detecting the others pri-miR listed in Supplementary information, Table S2. Primers used to detect target mRNA and miRNA machinery mRNA levels are listed in Supplementary information, Tables S3 and S4, respectively.
5′ rapid amplification of cleavage end and Nested PCR
RNA ligase-mediated rapid amplification of 5′ cDNA ends for analysis of target cleavage product was carried out according to GeneRacerII™ (RLM-RACE, Invitrogen) instruction manual. Complementary DNA was used as template in 5′ RACE PCR with primer specific to the RNA adapter sequence and a locus-specific reverse primer (Supplementary information, Table S5). Additional PCR reactions with nested primers (Supplementary information, Table S6) were performed for increase the specificity and sensitivity of RACE products.
Chromatin immunoprecipitations
ChIP assays were performed as described previously 42. Seeds of Ws and gcn5 were sterilized, kept for 2 days at 4 °C, and grown in vitro under long-day conditions. 12-day-old seedlings were harvested and fixed in 1% formaldehyde for 15 min in a vacuum and then neutralized by 0.125 M glycine. After washing with sterilized water, the samples were ground in liquid nitrogen as described above. Nuclei pellets were suspended in a buffer containing 0.25 M sucrose, 10 mM Tris-HCl, pH 8, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, 0.1 mM PMSF and protease inhibitors (one minitablet per milliliter; Roche). The suspensions were transferred to microfuge tubes and centrifuged at 12 000× g for 10 min. The pellets were suspended in 1.7 M sucrose, 10 mM Tris-HCl, pH 8, 2 mM MgCl2, 0.15% Triton X-100, 5 mM β-mercaptoethanol, 0.1 mM PMSF and protease inhibitors, and centrifuged through a layer of the same buffer in microfuge tubes. The nuclear pellets were lysed in a buffer containing 50 mM Tris-HCl, pH 8, 10 mM EDTA, 1% SDS and protease inhibitors. The lysed nuclei were sonicated four times for 15 s at 4 °C followed by centrifugation. The supernatants containing chromatin fragments were diluted 10-fold with 1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8 and 167 mM NaCl. Aliquots of the dilution were used for an immunoprecipitation assay. The antibody of GCN5 was described in Benhamed et al. 9. Immunoprecipitated DNA was analyzed by PCR using primer sets listed in Supplementary information, Table S2.
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
References
Berger SL . The complex language of chromatin regulation during transcription. Nature 2007; 447:407–412.
Grant PA, Duggan L, Cote J, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev 1997; 11:1640–1650.
Lee TI, Causton HC, Holstege FC, et al. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 2000; 405:701–704.
Carre C, Szymczak D, Pidoux J, Antoniewski C . The histone H3 acetylase dGcn5 is a key player in Drosophila melanogaster metamorphosis. Mol Cell Biol 2005; 25:8228–8238.
Xu W, Edmondson DG, Evrard YA, et al. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genet 2000; 26:229–232.
Yamauchi T, Yamauchi J, Kuwata T, et al. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc Natl Acad Sci USA 2000; 97:11303–11306.
Pandey R, Muller A, Napoli CA, et al. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res 2002; 30:5036–5055.
Vlachonasios KE, Thomashow MF, Triezenberg SJ . Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 2003; 15:626–638.
Benhamed M, Bertrand C, Servet C, Zhou DX . Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 2006; 18:2893–2903.
Bertrand C, Bergounioux C, Domenichini S, Delarue M, Zhou DX . Arabidopsis histone acetyltransferase AtGCN5 regulates the floral meristem activity through the WUSCHEL/AGAMOUS pathway. J Biol Chem 2003; 278:28246–28251.
Earley KW, Shook MS, Brower-Toland B, Hicks L, Pikaard CS . In vitro specificities of Arabidopsis co-activator histone acetyltransferases: implications for histone hyperacetylation in gene activation. Plant J 2007; 52:615–626.
Benhamed M, Martin-Magniette ML, Taconnat L, et al. Genome-scale Arabidopsis promoter array identifies targets of the histone acetyltransferase GCN5. Plant J 2008; 56:493–504.
Bertrand C, Benhamed M, Li YF, Ayadi M, et al. Arabidopsis HAF2 gene encoding TATA-binding protein (TBP)-associated factor TAF1, is required to integrate light signals to regulate gene expression and growth. J Biol Chem 2005; 280:1465–1473.
Park W, Li J, Song R, Messing J, Chen X . CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 2002; 12:1484–1495.
Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J . SERRATE: a new player on the plant microRNA scene. EMBO Rep 2006; 7:1052–1058.
Vazquez F, Gasciolli V, Crete P, Vaucheret H . The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol 2004; 14:346–351.
Boutet S, Vazquez F, Liu J, Beclin C, et al. Arabidopsis HEN1: a genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr Biol 2003; 13:843–848.
Vaucheret H, Vazquez F, Crete P, Bartel DP . The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 2004; 18:1187–1197.
Palatnik JF, Allen E, Wu X, et al. Control of leaf morphogenesis by microRNAs. Nature 2003; 425:257–263.
Chen X . A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 2004; 303:2022–2025.
Guo HS, Xie Q, Fei JF, Chua NH . MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 2005; 17:1376–1386.
Lauter N, Kampani A, Carlson S, Goebel M, Moose SP . microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci USA 2005; 102:9412–9417.
Jones-Rhoades MW, Bartel DP . Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 2004; 14:787–799.
Lu S, Sun YH, Shi R, et al. Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 2005; 17:2186–2203.
Sunkar R, Zhu JK . Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004; 16:2001–2019.
Zhou X, Wang G, Sutoh K, Zhu JK, Zhang W . Identification of cold-inducible microRNAs in plants by transcriptome analysis. Biochim Biophys Acta 2008; 1779:780–788.
Chiou TJ, Aung K, Lin SI, et al. Regulation of phosphate homeostasis by MicroRNA in Arabidopsis. Plant Cell 2006; 18:412–421.
Bari R, Datt Pant B, Stitt M, Scheible WR . PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 2006; 141:988–999.
Achard P, Herr A, Baulcombe DC, Harberd NP . Modulation of floral development by a gibberellin-regulated microRNA. Development 2004; 131:3357–3365.
Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK . A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 2005; 15:2038–2043.
Llave C, Xie Z, Kasschau KD, Carrington JC . Cleavage of scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 2002; 297:2053–2056.
Servet C, Benhamed M, Latrasse D, et al. Characterization of a phosphatase 2C protein as an interacting partner of the histone acetyltransferase GCN5 in Arabidopsis. Biochim Biophys Acta 2008; 1779:376–382.
Prigge MJ, Wagner DR . The Arabidopsis serrate gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 2001; 13:1263–1279.
Nikovics K, Blein T, Peaucelle A, et al. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 2006; 18:2929–2945.
Tian L, Chen ZJ . Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc Natl Acad Sci USA 2001; 98:200–205.
Parizotto EA, Dunoyer P, Rahm N, Himber C, Voinnet O . In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev 2004; 18:2237–2242.
Wang JW, Wang LJ, Mao YB, et al. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 2005; 17:2204–2216.
Zhang L, Volinia S, Bonome T, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci USA 2008; 105:7004–7009.
Scott GK, Mattie MD, Berger CE, Benz SC, Benz CC . Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res 2006; 66:1277–1281.
Grigg SP, Canales C, Hay A, Tsiantis M . SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature 2005; 437:1022–1026.
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR . Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 2005; 139:5–17.
Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA . Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 2002; 297:1871–1873.
Rhoades MW, Reinhart BJ, Lim LP, et al. Prediction of plant microRNA targets. Cell 2002; 110:513–520.
Schwab R, Palatnik JF, Riester M, et al. Specific effects of microRNAs on the plant transcriptome. Dev Cell 2005; 8:517–527.
Palatnik JF, Wollmann H, Schommer C, et al. Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev Cell 2007; 13:115–125.
Reyes JL, Chua NH . ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J 2007; 49:592–606.
Xie Z, Kasschau KD, Carrington JC . Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol 2003; 13:784–789.
Laufs P, Peaucelle A, Morin H, Traas J . MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 2004; 131:4311–4322.
Mallory AC, Dugas DV, Bartel DP, Bartel B . MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol 2004; 14:1035–1046.
Li H, Xu L, Wang H, et al. The Putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and MicroRNA165/166 in Arabidopsis leaf development. Plant Cell 2005; 17:2157–2171.
Mallory AC, Reinhart BJ, Jones-Rhoades MW, et al. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5' region. EMBO J 2004; 23:3356–3364.
Wu MF, Tian Q, Reed JW . Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 2006; 133:4211–4218.
Aukerman MJ, Sakai H . Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell Online 2003; 15:2730.
Jung JH, Seo YH, Seo PJ, et al. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 2007; 19:2736–2748.
Aung K, Lin SI, Wu CC, et al. pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol 2006; 141:1000–1011.
Lin SI, Chiang SF, Lin WY, et al. Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol 2008; 147:732–746.
Acknowledgements
This work was supported by grants from the French Centre National de la Recherche Scientifique (CNRS, France).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary information, Table S1
List of probe of miRNA for northern blots (PDF 9 kb)
Supplementary information, Figure S1
Detection of pri–miRNA levels in gcn5 in comparison with wild type by gel–based RT–PCR. (PDF 160 kb)
Supplementary information, Figure S2
Accumulation of different members of pri-miR 172, 395 and 399 gene families in inflorescence (left) and seedlings (right) of gcn5-2 and wild type plants. (PDF 69 kb)
Supplementary information, Figure S3
GCN5 is not associated with RNAi machinery genes. (PDF 91 kb)
Supplementary information, Table S2
List of primers for expression of pri-miRNA and for ChIP analysis by quantitative PCR (PDF 10 kb)
Supplementary information, Table S3
List of primers for miRNA target gene expression analysis by RT-PCR (PDF 10 kb)
Supplementary information, Table S4
List of primers for miRNA machinery genes used for RT-PCR (PDF 9 kb)
Supplementary information, Table S5
List of primers for RLM-RACE PCR (PDF 9 kb)
Supplementary information, Table S6
List of primers for Nested PCR (PDF 9 kb)
Rights and permissions
About this article
Cite this article
Kim, W., Benhamed, M., Servet, C. et al. Histone acetyltransferase GCN5 interferes with the miRNA pathway in Arabidopsis. Cell Res 19, 899–909 (2009). https://doi.org/10.1038/cr.2009.59
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2009.59
Keywords
This article is cited by
-
Micro RNA mediated regulation of nutrient response in plants: the case of nitrogen
Plant Physiology Reports (2022)
-
Exploring the Agrobacterium-mediated transformation with CRISPR/Cas9 in cucumber (Cucumis sativus L.)
Molecular Biology Reports (2022)
-
Regulation of pri-MIRNA processing: mechanistic insights into the miRNA homeostasis in plant
Plant Cell Reports (2021)
-
HAG1 and SWI3A/B control of male germ line development in P. patens suggests conservation of epigenetic reproductive control across land plants
Plant Reproduction (2021)
-
Transcription factor TERF1 regulates nuclear genes expression through miRNAs in tobacco under drought stress condition
Plant Growth Regulation (2019)