Abstract
Arabidopsis thaliana leucine-rich repeat-containing (NLR) proteins RPS4 and RRS1, known as dual resistance proteins, confer resistance to multiple pathogen isolates, such as the bacterial pathogens Pseudomonas syringae and Ralstonia solanacearum and the fungal pathogen Colletotrichum higginsianum. RPS4 is a typical Toll/interleukin 1 Receptor (TIR)-type NLR, whereas RRS1 is an atypical TIR-NLR that contains a leucine zipper (LZ) motif and a C-terminal WRKY domain. RPS4 and RRS1 are localised near each other in a head-to-head orientation. In this study, direct mutagenesis of the C-terminal LZ motif in RRS1 caused an autoimmune response and stunting in the mutant. Co-immunoprecipitation analysis indicated that full-length RPS4 and RRS1 are physically associated with one another. Furthermore, virus-induced gene silencing experiments showed that hypersensitive-like cell death triggered by RPS4/LZ motif-mutated RRS1 depends on EDS1. In conclusion, we suggest that the RRS1-LZ motif is crucial for the regulation of the RPS4/RRS1 complex.
Similar content being viewed by others
Introduction
In both plants and animals, intracellular immune receptors of the nucleotide-binding domain and leucine-rich repeat (NLR) protein superfamily play an important role in pathogen recognition and effective innate immune responses1,2. Plant NLRs use either a direct or an indirect mode for pathogen effector recognition; they either directly bind the cognate effectors or detect changes in host proteins caused by the cognate effectors and subsequently trigger plant defence responses. These modes are known as effector triggered immunity (ETI), which is usually associated with a hypersensitive reaction that often includes localised cell death3.
Plant NLRs generally consist of a C-terminal leucine-rich repeat (LRR) domain and a central nucleotide-binding adaptor shared with Apaf-1, plant resistance proteins and CED-4 (NB-ARC)3. NLRs are diverse in their N-terminal structures and possess either a Toll/interleukin 1 receptor (TIR) domain or a coiled-coil (CC) domain4. As highly polymorphic and variable parts of plant NLRs5, the LRR domains are involved in protein–protein interaction and play a key role in effector recognition specificity6,7. In contrast, the NB-ARC domain is highly conserved among the majority of plant NLRs. When NLRs are inactive, the NB-ARC domain physically interacts with the LRR domain8,9,10. After effector recognition, the conformational change allows nucleotide exchanges, the replacement of ADP by ATP in the NB domain and subsequent NLR activation11.
Both the N-terminal TIR and CC domains of plant NLRs are considered directly connected to downstream defence signalling components. Overexpression of the TIR and CC domains from plant NLRs causes effector-independent hypersensitive (HR) cell death12,13. Crystal structure analysis of flax L6 and barley MLA10 also showed that the homodimerisation of the TIR and CC domains is necessary for downstream defence signalling activity12,13. In addition, the N-terminal domains of plant NLRs may also participate in effector recognition.
Plant NLRs are capable of self-association in the pre- or post-activation state of NLR complexes1. However, some plant NLRs require a second NLR for defence signalling14 and they form heterodimers or hetero-oligomers. Recent reports indicated that some NLR pairs are required for full disease resistance14,15. In our previous studies, Arabidopsis thaliana NLRs RPS4 (Resistance to Pseudomonas syringae 4) and RRS1 (Resistance to Ralstonia solanacearum 1), known as dual resistance proteins, confer resistance to multiple pathogen isolates, such as the bacterial pathogens P. syringae and R. solanacearum and the fungal pathogen Colletotrichum higginsianum16. RPS4 is a typical TIR-type NLR, whereas RRS1 is an atypical TIR-NLR that contains a leucine zipper (LZ) motif and a C-terminal WRKY domain. RPS4 and RRS1 are localised in near each other in a head-to-head orientation. This NLR pair is required to recognise the AvrRps4 effector protein from P. syringae, the PopP2 effector protein from R. solanacearum and one or more unidentified effectors from C. higginsianum. We recently demonstrated that the introduction of both RPS4 and RRS1 confers resistance to multiple, taxonomically distinct pathogen families17,18. The successful transfer of the R gene pair to some plant families (i.e., Brassicaceae, Solanaceae and Cucurbitaceae) implies that the downstream components of the NLR pair are highly conserved.
The precise mechanism of how RPS4 functions with RRS1 is not clear. Both RPS4 and RRS1 are partially localised in the nucleus of living plant cells in the presence of effectors19,20. The two proteins likely form a heterodimer and assemble into a complex. Previous studies suggested that RPS4-TIR physically interacts with RRS1-TIR21 and the RPS4/RRS1 complex enables the perception of pathogen effectors, AvrRps4 and PopP2, which target the RRS1 WRKY domain and activate defence responses22,23. RPS4 is also physically associated with EDS124,25. This TIR-domain heterodimerisation plays a role in the effector recognition complex consisting of RPS4/RRS1. The objectives of this study were to investigate whether: 1) the full-length RPS4 is physically associated with RRS1 and 2) the RRS1-LZ motif plays a role in the regulation of RPS4/RRS1.
Results
Transient expression of dual R proteins
To investigate the expression of dual R proteins, RPS4 and RRS1, in Nicotiana benthamiana leaves using Agrobacterium-mediated transient expression assays, 3 × FLAG and 4 × Myc tags were fused into the N-terminus of RPS4 and RRS1, respectively. These tagged constructs were driven by the cauliflower mosaic virus 35S promoter, while the omega leader sequence, which is known to act as a translational enhancer in plants, served as the 5′ UTR26,27 (Fig. 1a).
Transient expression of dual R proteins.
(a) Epitope-tagged constructs used in the transient expression. (b) Immunodetection of microsomal extracts from Nicotiana benthamiana leaves transiently expressed FLAG-RPS4 and/or Myc-RRS1 44 h after Agrobacterium infiltration. (c) Subcellular localisation of FLAG-RPS4, Myc-RRS1 and Myc-RRS1Δlz with immunoblotting. Transiently co-expressed FLAG-RPS4 and Myc-RRS1 or Myc-RRS1Δlz mainly localised in microsomal fractions 44 h after Agrobacterium infiltration. Microsomal (Micro) and nuclear (Nucl) fractions are 10- and 60-fold more concentrated, respectively, compared with the soluble (Sol) fraction. The degree of fraction enrichment was determined using antibodies against marker proteins (cytoplasmic soluble; anti-GAPDH, microsomes; anti BiP and nucleus; anti-Histone H3).
Immunoblot assays of transiently expressed or co-expressed FLAG-RPS4 and Myc-RRS1 in N. benthamiana showed that co-expressed full-length RPS4 and RRS1 gave a strong signal on immunoblot compared with transiently expressed RRS1 or RPS4 that were both weakly detected (Fig. 1b). In addition, transiently co-expressed FLAG-RPS4 and Myc-RRS1 were mainly localised in N. benthamiana microsomal fractions and weakly localised in nucleus fractions (Fig. 1c). Several previous studies also showed that RPS4 was localised in the microsomal fraction25,28 and that when transiently expressed in N. benthamiana, RPS4 and SNC1 formed a common protein complex in cytoplasmic microsomal compartments28.
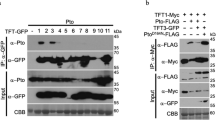
Co-immunoprecipitation (Co-IP) analysis on full-length RPS4 and RRS1 proteins isolated 44 h after Agrobacterium infiltration showed that they interacted with one another in the microsomal fraction (Fig. 2). The interaction between the RPS4 TIR domain and full-length RRS1 was not detected in the microsomal fraction with Co-IP, although both were detected in this fraction with immunoblotting. Our results suggested that other domains were also required for RPS4/RRS1 interaction, even though the TIR domains of RPS4 and RRS1 formed a heterodimer21.
Co-immunoprecipitation analysis of RPS4 and RRS1.
RPS4 and RRS1 were transiently expressed in Nicotiana benthamiana leaves induced by Agrobacterium infiltration for 44 h. Immunoprecipitation (IP) analyses of full-length or TIR domain RPS4 and full-length RRS1 were performed in microsomal fractions with the indicated antibody. The immunoprecipitates were immunoblotted (IB) with the indicated antibody.
To test whether tag-fused RPS4 and RRS1 confer resistance against C. higginsianum, we complementarily introduced N- or C-terminal HA-tagged RRS1 and N-terminal Myc-tagged RRS1 into the rrs1-1 A. thaliana Ws-2 accession mutant line. Resistance to C. higginsianum was fully restored in the transgenic plants with N-terminal tagged RRS1, but was not restored in the transgenic plants with C-terminal tagged RRS1 (Fig. S1). These results suggested that a proper C-terminus structure in RRS1 is important for C. higginsianum resistance in Arabidopsis. Similarly, we complementarily introduced N-terminal FLAG-tagged RPS4 and C-terminal YFP-tagged RPS4 into the rps4-21 A. thaliana Ws-2 accession mutant line. Resistance to C. higginsianum was fully restored in the transgenic plants with N-terminal tagged RPS4, but was not restored in the transgenic plants with C-terminal tagged RPS4 (Fig. S1). Thus, a proper C-terminus structure in RPS4 is likely also important for C. higginsianum resistance in Arabidopsis. In addition, we showed via immunoblotting that N-terminal tagged 4 × Myc-RRS1 and 3 × FLAG-RPS4 were detected in transgenic Arabidopsis, but the transgenic plants with C-terminal tagged RRS1-3 × HA, RPS4-YFP and N-terminal tagged 3 × HA-RRS1 were not detected (Fig. S1). The amount of these proteins may be undetectable with immunoblotting under our conditions. It is important that the N-terminal tagged RRS1 and RPS4 used are functional.
Mutations in the leucine zipper motif of RRS1
RPS4 is a typical TIR-type NLR, while RRS1 is a TIR-type NLR with original structure that contains a C-terminal WRKY domain, which is a class of DNA-binding transcription factors in plants29. In the present study, we found a LZ motif, LRVSYDDLQEMDKVLFLYIASL, located between the LRR and WRKY domains in RRS1 (Fig. 3a). We investigated the natural sequence variation of amino acid residues across 19 A. thaliana accessions for which large-scale single nucleotide polymorphism (SNP) genotyping of RRS1 is possible: Bay-0, Br-0, Bur-0, C24, Col-0, Cvi-0, Est-1, Fei-0, Ler-1, Lov-5, Nfa-8, Rrs-7, Rrs-10, Sha, Tamm-2, Ts-1, Tsu-1, Van-0 and Ws-230.
Analysis of leucine zipper (LZ) motif in RRS1.
(a) The predicted domains and motifs in RRS1 (Ws-2 accession). Schematic gene structure of RRS1 with exons shown as boxes and introns as lines. Localisation and amino acid positions of TIR, NB-ARC (nucleotide-binding adaptor shared with Apaf-1, plant resistance proteins and CED-4), leucine rich repeat (LRR), LZ, nuclear localisation signal (NLS) and WRKY domains is indicated. The transformation of Arabidopsis mutants was performed using the 8.2-kbp genomic RRS1 fragments (from the Ws-2 accession) with or without the LZ mutation. (b) RRS1Δlz mutant was generated by L to A mutations in LZ motif. (c) Phenotypes of 4-week-old wild-type Ws-2, mutants and transgenic plants under normal growth conditions. All mutants and transgenic plants originated from the Ws-2 accession.
The LZ motif is completely conserved in these 19 accessions, which contain both RRS1-R and RRS1-S. Saucet et al.31 had reported that RRS1B and RPS4B, similar to RRS1 and RPS4, confer recognition of AvrRps4 but not PopP2. In RRS1B from Ws-2, we also found two LZ motifs, LKGSLSSLPNVLRLLHWENYPL and LRVRYAGLQEIYKALFLYIAGL.
The LZ is a protein–protein interaction domain consisting of an α-helical conformation with a leucine residue at every seventh position, which often facilitates dimerisation32. To investigate the role of the LZ motif in RRS1 (from Ws-2), we generated mutations in three leucine residues within the zipper (Fig. 3b) and named the mutated protein RRS1Δlz. We observed that the rrs1-1/RRS1Δlz mutants grew abnormally and constitutively expressed the inducible defence gene, PR1. Compared with expression in wild-type Ws-2, PR1 gene expression was three hundred thousand- (rrs1-1/RRS1Δlz#1) and one hundred fifty thousand-fold (rrs1-1/RRS1Δlz#2) higher. Therefore, introducing RRS1Δlz induced autoimmune response (Figs 3c and 4). Interestingly, the rrs1-1/rps4-21/RRS1Δlz mutants, in which RPS4 was absent, grew normally and did not constitutively express PR1, suggesting that RPS4 was required for autoimmune response in the rrs1-1/RRS1Δlz mutant (Figs 3 and 4).
Expression of defence-related PR1 in transgenic plants under normal growth conditions.
Expression levels of PR1 in transgenic plants under normal growth conditions were monitored by quantitative real-time PCR. The relative expression level was normalised against the expression level of CBP20, which is constitutively expressed. Each sample was repeated at least three times. Bars indicate the standard error (SE). The asterisks indicate significant differences compared with wild-type Ws-2 (Dunnett’s method, P < 0.05)50. The nucleotide sequence of the gene-specific primer is listed in Table S1.
Transient expression of dual R proteins induces an HR-like cell death in N. benthamiana
Transient expression of both full-length RPS4 and RRS1Δlz induced HR-like cell death (Fig. 5a). The HR symptoms appeared 3 d after the injection of full-length RPS4 and RRS1Δlz into N. benthamiana. However, transient expression of full-length RRS1, RRS1Δlz, or RPS4/RRS1 did not cause HR-like cell death (Fig. 5a). In addition, we used immunoblotting to verify that HR absence was not due to absence of the R proteins. Co-expressed RRS1/RPS4 and RRS1Δlz/RPS4 yielded strong and weak signals on immunoblots, respectively (Fig. 5b). Moreover, RRS1Δlz and RRS1 were weakly detected in the total protein extraction from RRS1Δlz- and RRS1-injected N. benthamiana (Fig. 5b). These results confirm that the lack of HR was not due to the absence of R proteins.
Transient expression of dual R proteins induces hypersensitive (HR)-like cell death in Nicotiana benthamiana.
Leaves of N. benthamiana were infiltrated with Agrobacterium carrying full-length FLAG-gRPS4, Myc-gRRS1 and Myc-gRRS1Δlz. (a) HR-like cell death was observed at 3 d post-inoculation (dpi). The photograph was taken at 7 dpi under white light to visualise cell death. A set of gRPS4 and gRRS1Δlz induced HR-like cell death. (b) Immunodetection of non-mutated RPS4, as well as RRS1 with a mutated LZ motif. Transiently co-expressed FLAG-RPS4 and Myc-RRS1 or Myc-RRS1Δlz were detected in the total protein extraction from N. benthamiana 44 h after Agrobacterium infiltration. Immunodetection was performed with either anti-FLAG or anti-Myc antibodies.
Immunoprecipitation analysis of RPS4 and RRS1Δlz
Tissue samples of N. benthamiana were harvested 44 h after Agrobacterium infiltration. The nucleus and microsomal fractions were extracted and immunoassayed with anti-FLAG and anti-Myc antibodies. Under these conditions, FLAG-RPS4 and Myc-RRS1 were detected as strong signals on immunoblotting in the microsomal fraction and as weak signals in the nucleus fraction (Fig. 1c). In contrast, RPS4 and RRS1Δlz isolated 44 h after Agrobacterium infiltration were weakly detected in the microsomal fraction but not in the nucleus fraction, although both mRNAs were detected in the leaves (Fig. 1c and S2).
RPS4/RRS1Δlz-triggered cell death depends on EDS1
To investigate whether RPS4/RRS1Δlz-triggered cell death depends on EDS1, virus-induced gene silencing (VIGS) was used33. An N. benthamiana seedling was silenced for NbEDS1 by inoculation with Agrobacterium-carrying tobacco rattle virus TRV:EDS1. The N. benthamiana plants that were inoculated with Agrobacterium harbouring TRV:GFP were used as control. Following the first inoculation, RPS4 and RRS1Δlz were transiently expressed in the upper leaves of silenced N. benthamiana plants. The HR-like cell death phenotype was monitored at 7 d post-inoculation (dpi). The results indicated that co-expression of RPS4 and RRS1Δlz conferred an HR-like cell death in the TRV:00 silenced control plants (Fig. 6a). Silencing of NbEDS1 in N. benthamiana completely abolished the HR phenotype (Fig. 6a). This result indicated that VIGS of NbEDS1 impaired HR because of RPS4 and RRS1Δlz co-expression and that NbEDS1 was required for RPS4/RRS1Δlz-triggered HR in N. benthamiana. Using VIGS, we found that cell death resulting from the co-expression of RPS4 and RRS1Δlz was due to the signalling component EDS1.
EDS1 is required for dual R protein-induced hypersensitive (HR)-like cell death.
TRV:Vector or TRV:EDS1-silenced N. benthamiana leaves were infiltrated with Agrobacterium containing FLAG-gRPS4 and Myc-gRRS1 or Myc-gRRS1Δlz constructs. (a) HR-like cell death was observed at 3 dpi. The photograph was taken at 7 dpi. (b) Immunodetection of non-mutated RPS4, as well as RRS1 with a mutated LZ motif. Total proteins extracted from N. benthamiana leaves 44 h after Agrobacterium infiltration showed the transient co-expression of FLAG-RPS4 and Myc-RRS1 or Myc-RRS1Δlz. Immunodetection was performed with either anti-FLAG or anti-Myc antibodies.
In addition, we used immunoblotting of the total protein extraction to analyse RRS1 and RRS1Δlz accumulation in both the presence and absence of RPS4 and EDS1 (Fig. 6b). With RPS4/RRS1 co-expressed N. benthamiana, EDS1 absence resulted in a decrease of RRS1 and RPS4 accumulation, compared with EDS1 presence. However, RRS1Δlz was weakly detected in both the presence and absence of EDS1. Finally, with RRS1Δlz expressed N. benthamiana, RRS1Δlz was weakly detected in the absence of RPS4.
VIGS is known to reduce the level of target mRNA34,35. To investigate whether TRV:EDS1 silencing causes a reduction in NbEDS1 mRNA, we performed mRNA expression analysis. NbEDS1 mRNA drastically decreased in TRV:EDS1-silenced plants after the second inoculation (Fig. S3). However, NbEDS1 mRNA was expressed in the control plants infected with TRV:GFP after the second inoculation (Fig. S3).
Characterisation of RPS4/RRS1Δlz-triggered cell death
Nuclear localisation of RPS4, containing nuclear localisation sequence (NLS), is necessary for AvrRps4-triggered cell death19,36. We investigated whether HR was abolished when RRS1Δlz was co-expressed with RPS4Δnls. We found that RRS1Δlz/RPS4Δnls did not induce HR (Fig. 7). Therefore, nuclear localisation of RPS4 is necessary for RRS1Δlz-mediated cell death.
Characterisation of RPS4/RRS1Δlz-triggered cell death in Nicotiana tabacum.
Leaves of N. tabacum were infiltrated with Agrobacterium carrying full-length FLAG-gRPS4, FLAG-gRPS4Δnls, Myc-gRRS1 and Myc-gRRS1Δlz. HR-like cell death was observed at 2 dpi, induced by sets of gRPS4/gRRS1Δlz and gRPS4/gRRS1/gRRS1Δlz. The photograph was taken at 4 dpi under white light to visualise cell death.
Co-expression of RRS1WT was reported to prevent RRS1-SLH1/RPS4-dependent constitutive HR, indicating that RRS1-SLH1-dependent auto-activation is recessive36. We also investigated whether RRS1Δlz/RPS4-dependent constitutive HR was prevented by co-expression of RRS1WT. Our results revealed that HR was weakened, but not abolished, when triggered by RRS1Δlz/RPS4/RRS1WT as opposed to RRS1Δlz/RPS4. These data suggest that RRS1WT did not completely interfere with RRS1Δlz/RPS4 triggered HR (Fig. 7). Therefore, auto-active alleles of RRS1Δlz are semi-dominant.
Discussion
Dual R proteins, RPS4 and RRS1, function as a complex and mediate defence response16,17. Zhang and Gassmann37 previously reported that 35S:FLAG-genomicRPS4-Ler construct produced low levels of full-length protein in N. benthamiana. In this study, we showed that full-length N-terminally FLAG-tagged RPS4 was only slightly detected in the microsomal fraction eluted from N. benthamiana leaves in the absence of RRS1. In contrast, co-expressed full-length RPS4 and RRS1 gave strong signals on immunoblots, indicating that RPS4/RRS1 is stable in planta. Using co-immunoprecipitation assays, we found that full-length RPS4 and RRS1 strongly interact with one another in planta. Williams et al. 2014 also showed that RPS4 interacts with the RRS1 TIR domain21. However, co-expressed full-length RPS4 and RRS1Δlz were weakly detected in the microsomal fraction and the total protein extraction eluted from N. benthamiana leaves. Additionally, we verified with quantitative real-time polymerase chain reaction (qRT-PCR) that RPS4 and RRS1 mRNAs accumulated at similar levels in N. benthamiana leaves (Fig. S2).
Previous studies showed that the N-terminal NBS and LZ motif are critical for the function of NBS-LRR type R protein, RPS238 and that WRKY transcription factors play an important role in plant defence39. WRKY 18, WRKY 40 and WRKY60 have potential LZ motifs at the N-terminus that are involved in the physical interaction of these WRKY proteins40. WRKYs with no LZ motifs are unable to interact with themselves and with each other. The C-terminal portion of RRS1 also possesses two conspicuous structural motifs, the LZ and the WRKY domain, that play a key role in protein–protein interaction. In this study, direct mutagenesis revealed that the C-terminal LZ motif in RRS1 is important for RPS4/RRS1 immunity. Noutoshi et al. also showed that a 3-bp insertion mutation of the WRKY domain in RRS1 (named RRS1-SLH1) causes autoimmunity41.
In this study, transient co-expression of RPS4/RRS1Δlz in N. tabacum and N. benthamiana induced strong HR-like cell death. However, we did not observe HR-like cell death in N. benthamiana induced by the transient expression of full-length RRS1, RRS1Δlz, or RPS4/RRS1. In N. tabacum, when transient RPS4 expression in the absence of AvrRps4 causes HR, it is called the R gene overdose effect19,42. We also observed stunting and mimic lesions in RPS4/RRS1Δlz-transgenic Arabidopsis plants, but not in rps4/RRS1Δlz transgenic plants without RPS4. We assume that RRS1 regulates the activation of RPS4 in the absence of Avr proteins and Avr proteins either direct or indirect modify RRS1 to activate RPS4. The structural change of RRS1 by Avr proteins is necessary for the activation of downstream defence responses. Therefore, the coexistence of RRS1 and RPS4 is essential for pathogen recognition and defence responses. On the other hand, RRS1B and RPS4B also confer recognition of AvrRps4 but not PopP231. In our preliminary experiments, we found that RRS1B was not required for resistance to C. higginsianum.
Interestingly, we found that Arabidopsis mutants complemented by C-terminally tagged RPS4- or RRS1-transgenes did not confer resistance to C. higginsianum, whereas those complemented by N-terminally tagged RPS4- or RRS1-transgenes did. These results imply that the C-termini of both RPS4 and RRS1 play a key role in the activation of RPS4/RRS1-mediated defence responses. We previously reported that RRS1 alleles from three C. higginsianum-susceptible accessions, Bur-0, Col-0 and Cvi-0, contain a premature stop codon16. In these accessions, the C-terminal region of RRS1 that follows the WRKY domain is relatively short compared with several resistant accessions (Ws-2 etc.). Recent reports22,23 suggest that C-terminal extension is necessary for resistance signalling, because the C-terminal region is required for PopP2 but not for AvrRps4. Thus, the C-terminal region of RRS1 likely plays a key role in the activation of RPS4/RRS1-mediated defence responses.
EDS1 encodes a lipase-like protein that functions in R protein-mediated and basal plant disease resistance43. In addition, EDS1 and NbEDS1 are required for RPS4-mediated disease resistance in Arabidopsis and tobacco, respectively42,44. In present study, we found that RPS4/RRS1Δlz-triggered HR is completely abolished in NbEDS1-silenced N. benthamiana plants. Our results suggested that RPS4/RRS1Δlz-triggered HR also requires NbEDS1 as a signalling component in N. benthamiana plants.
In conclusion, our data indicated that RPS4/RRS1 is required for active defence responses and some structural domains that constitute these R proteins contribute to the interaction of RPS4 with RRS1.
Methods
Plant materials
A. thaliana plants were grown in soil mix (Sakata Seed Corp.) and expanded vermiculite (2–5 mm granules) at a 1:1 ratio, in a growth chamber at 22 °C under a medium-day photoperiod (12-h light/12-h night). The accessions, mutants and transformants used in this study are of Ws-2 background. Tobacco plants (N. benthamiana and N. tabacum) were grown in soil mix and expanded vermiculite (2–5 mm granules) at a 2:1 ratio, in a growth chamber at 25 °C under a long-day photoperiod (16-h light/8-h night).
Construction of the R-gene plasmid
All DNA fragments containing RRS1 and/or RPS4 used in this study derived from genome of the A. thaliana Ws-2 accession. Plasmids used in this study were constructed by Gateway® technology following manufacturer protocol (Life Technologies, USA). All clones were verified with DNA sequencing. The pCR8GW-RR-Ws plasmid was cloned using a 10.9-kbp genomic fragment containing RPS4 and RRS1 as previously described17. The genomic fragment of RRS1 (coding region) was PCR-amplified using pCR8GW-RR-Ws as template and cloned into pCR8GW-TOPO (named pCR8GW-gRRS1). To generate the destination vector pGWB18Ω (35S:Ω:4xMyc), the HindIII and XbaI regions in pGWB18 were replaced with 35S enhancers and the omega leader sequence cassette in pBE211345. To create the destination vector pGWB18-RRS1p, the 35S promoter region in pGWB18 was replaced with the RRS1 promoter region (1.8 kbp upstream of the start codon) using the In-Fusion® HD Cloning Kit (Takara Bio Inc., Japan). To generate the 35S:Ω:4 × Myc-gRRS1 and RRS1p:4 × Myc-gRRS1 constructs, LR reactions were performed to recombine the entry clone, containing genomic RRS1, into the Gateway®-compatible destination vectors: pGWB18Ω for Agrobacterium-mediated transient expression and pGWB18-RRS1p for stable Arabidopsis transformation via LR reaction. Site-directed mutagenesis of genomic RRS1 with or without the promoter region16 was performed by a custom cloning service (Takara Bio Inc., Japan) to generate RRS1Δlz carrying Leu to Ala mutations (L1089A, L1096A and L1103A) at the LZ motif. Subsequently, LR cloning was used to generate binary constructs in pGWB18Ω for Agrobacterium-mediated transient expression and in pGWB1 for stable Arabidopsis transformation.
The 6.3-kbp genomic RPS4 fragment, including approximately 2.1-kbp upstream and 109-bp downstream regions, was cloned into pCR8GW-TOPO (named pCR8GW- RPS4p:gRPS4). To generate the RPS4p:3 × FLAG-gRPS4 construct, synthetic 3 × FLAG and a spacer sequence, MDYKDHDGDYKDHDIDYKDDDDKGGGS, was inserted just before the RPS4 start codon by a custom cloning service (Life Technologies, USA). To generate the pCR8GW-35S:Ω:3 × FLAG-gRPS4, the RPS4 promoter region in pCR8GW-RPS4p:gRPS4 was replaced by 35S enhancers and the omega leader sequence cassette in pBE2113, using the In-Fusion® HD Cloning Kit (Takara Bio Inc., Japan). To create the destination vector pBGYNΔEYFP-nuc, the EYFP-nuc cassette in pBGYN (Inplanta Innovations Inc., Japan) was cut with SmaI and SacI. The attR2 region in the Gateway® cassette was engineered with PCR and inserted into the vector using the In-Fusion® HD Cloning Kit (Takara Bio Inc., Japan). RPS4p:3 × FLAG-gRPS4 and 35S:Ω:3 × FLAG-gRPS4 were recombined into the destination vectors, pBGYNΔEYFP-nuc, for stable Arabidopsis transformation and pGWB1 for Agrobacterium-mediated transient expression via LR reaction. The DNA fragment of 35S: Ω:3 × FLAG-gRPS4-TIR (encoding the N-terminal amino acids 1–235) was PCR-amplified using pGWB1-35S:Ω:3 × FLAG-gRPS4 as a template and cloned into pCR8GW-TOPO. The 35S:Ω:3 × FLAG-gRPS4-TIR construct was sub-cloned into the destination vector pGWB1 via LR reaction.
Arabidopsis transformation
Arabidopsis transformation was carried out according to the floral inoculating method using Agrobacterium tumefaciens strain GV3101 (pMP90)46.
C. higginsianum inoculation and quantification of C. higginsianum actin mRNA
C. higginsianum Saccardo isolates (MAFF305635) were obtained from the Ministry of Agriculture, Forestry and Fisheries (MAFF) Genebank, Japan. Arabidopsis plants were inoculated as described previously47 and harvested at 5 dpi for qRT-PCR analysis. The quantification of C. higginsianum was performed as described previously47.
Transient expression assay in N. benthamiana and N. tabacum
N. benthamiana and N. tabacum plants were grown in a growth chamber at 25 °C under a long-day photoperiod (16-h light/8-h night). Nicotiana benthamiana (32-day-old) and N. tabacum (32-day-old) plants were used in the experiment. Overnight bacterial cultures of A. tumefaciens strain GV3101 (pMP90) containing constructs with epitope-tags were harvested with centrifugation. Cells were washed three times in an induction buffer (10 mM 2-(N-morpholino)ethanesulfonic acid, pH 5.6, 10 mM MgCl2 and 150 μM acetosyringone), re-suspended in an induction buffer and incubated for 2–4 h in darkness at 25 °C. For transient expression experiments, bacterial strains containing the constructs were mixed, adjusting each strain concentration to a final optical density of 0.5 at 600 nm (OD600). Agrobacterium cells were hand-infiltrated into N. benthamiana and N. tabacum leaves with a 1-mL blunt-end syringe. Cell death responses began to appear on leaves 3 and 2 d after Agrobacterium infiltration in N. benthamiana and N. tabacum, respectively. Symptoms were documented under white light to visualise cell death.
Protein fractionation and immunoblotting
Microsomal and soluble fractions were prepared as described previously28. Extraction buffer (50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.5, 250 mM sucrose, 15 mM ethylenediaminetetraacetic acid, 5% glycerol, 0.5% polyvinylpyrrolidone, 3 mM dithiothreitol, 2× Roche protease-inhibitor cocktail and 1× Roche phosphatase-inhibitor cocktail) (Roche, Switzerland) was added to the plant materials, which was ground with mortar and pestle, then with a Potter-Elvehjem tissue grinder. The extracts were filtered through two layers of miracloth pre-wetted with extraction buffer and centrifuged at 2,000 × g for 10 min at 4 °C. The supernatant consisting of the cytoplasmic fraction was further subjected to ultracentrifugation at 100,000 × g to separate the soluble and microsomal (pellet) fractions. The pellet was resuspended in extraction buffer containing 0.5% Igepal CA-630 (Sigma-Aldrich, USA).
Nuclear extracts were prepared with a semi-pure preparation method using the CelLyticTM PN Isolation/Extraction Kit (Sigma-Aldrich, USA) and following manufacturer protocol, with one modification: 1× Roche phosphatase-inhibitor cocktail was added into NIBA buffer.
Total proteins were extracted according to previously described methods23.
Total protein was separated on a 4–15% sodium dodecyl sulfate polyacrylamide gel (BioRad, USA) and transferred onto a polyvinylidene difluoride membrane. Immunoblots were performed with monoclonal anti-FLAG (Sigma-Aldrich, USA) and anti-c-Myc (Roche, Switzerland) antibodies, as well as a secondary HRP-conjugated anti-mouse antibody (Promega, USA), then visualised with chemiluminescence (ECL; Bio-Rad, USA). The degree of enrichment in cellular fractionation was determined by immunoblot analyses with anti-GAPDH (Genscript, USA), anti-BiP (Agrisera, Sweden) and anti-histone H3 (Abcam, USA) antibodies. Secondary HRP-conjugated anti-mouse, anti-goat and anti-rabbit antibodies (Promega, USA) were used for detecting chemiluminescent immunoblots.
The co-immunoprecipitation assays were performed using Anti-FLAG M2 affinity and anti-c-Myc affinity gels (Sigma, USA) following manufacturer protocol. The protein concentrations of fractions were determined with Bradford assays48 using bovine serum albumin as a standard. Protein samples (1 mg) of microsomal fractions were incubated with Anti-FLAG M2 for 2 h or anti-c-Myc for 1 h affinity gels (Sigma-Aldrich, USA) at 4 °C. The immunoprecipitates were analysed with immunoblotting using monoclonal anti-FLAG (Sigma-Aldrich, USA) and anti-c-Myc (Roche, Switzerland) antibodies. A special secondary antibody (Mouse TrueBlot anti-Mouse Ig HRP; Rockland, USA) that only detects full-length immunoglobulin was used for immunoblotting to avoid any nonspecific bands.
VIGS
Using a tobacco rattle virus vector, VIGS of the N. benthamiana homologue EDS1 was performed as described previously49. Nicotiana benthamiana plants (14-day-old) were infiltrated with Agrobacteria carrying TRV:EDS1 to silence NbEDS1. After 3 to 4 weeks, the upper leaves of these plants were used for an Agrobacterium-mediated transient assay.
Additional Information
How to cite this article: Narusaka, M. et al. Leucine zipper motif in RRS1 is crucial for the regulation of Arabidopsis dual resistance protein complex RPS4/RRS1. Sci. Rep. 6, 18702; doi: 10.1038/srep18702 (2016).
References
Bonardi, V. & Dangl, J. L. How complex are intracellular immune receptor signaling complexes? Front. Plant Sci. 3, 237 (2012).
Jones, J. D. & Dangl, J. L. The plant immune system. Nature 444, 323–329 (2006).
van der Biezen, E. A. & Jones, J. D. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 23, 454–456 (1998).
Meyers, B. C., Kozik, A., Griego, A., Kuang, H. & Michelmore, R. W. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15, 809–834 (2003).
Bakker, E. G., Toomajian, C., Kreitman, M. & Bergelson, J. A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell 18, 1803–1818 (2006).
Farnham, G. & Baulcombe, D. C. Artificial evolution extends the spectrum of viruses that are targeted by a disease-resistance gene from potato. Proc. Natl. Acad. Sci. USA 103, 18828–18833 (2006).
Sela, H. et al. Ancient diversity of splicing motifs and protein surfaces in the wild emmer wheat (Triticum dicoccoides) LR10 coiled coil (CC) and leucine-rich repeat (LRR) domains. Mol. Plant Pathol. 13, 276–287 (2012).
Moffett, P., Farnham, G., Peart, J. & Baulcombe, D. C. Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21, 4511–4519 (2002).
Leister, R. T. et al. Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell 17, 1268–1278 (2005).
Ade, J., DeYoung, B. J., Golstein, C. & Innes, R. W. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc. Natl. Acad. Sci. USA 104, 2531–2536 (2007).
Lukasik, E. & Takken, F. L. W. STANDing strong, resistance proteins instigators of plant defence. Curr. Opin. Plant Biol. 12, 427–436 (2009).
Bernoux, M. et al. Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling and autoregulation. Cell Host Microbe. 9, 200–211 (2011).
Maekawa, T. et al. Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe. 9, 187–199 (2011).
Eitas, T. K. & Dangl, J. L. NB-LRR proteins: pairs, pieces, perception, partners and pathways. Curr. Opin. Plant Biol. 13, 472–477 (2010).
Okuyama, Y. et al. A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J. 66, 467–479 (2011).
Narusaka, M. et al. RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 60, 218–226 (2009).
Narusaka, M. et al. Interfamily transfer of dual NB-LRR genes confers resistance to multiple pathogens. PLoS One 8, e55954 (2013).
Narusaka, M., Hatakeyama, K., Shirasu, K. & Narusaka, Y. Arabidopsis dual resistance proteins, both RPS4 and RRS1, are required for resistance to bacterial wilt in transgenic Brassica crops. Plant Signal. Behav. 9, e29130 (2014).
Wirthmueller, L., Zhang, Y., Jones, J. D. & Parker, J. E. Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 17, 2023–2029 (2007).
Deslandes, L. et al. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA 100, 8024–8029 (2003).
Williams, S. J. et al. Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344, 299–303 (2014).
Roux, C. L. et al. A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 161, 1074–1088 (2015).
Sarris, P. F. et al. A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161, 1089–1100 (2015).
Heidrich, K. et al. Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science 334, 1401–1404 (2011).
Bhattacharjee, S., Halane, M. K., Kim, S. H. & Gassmann, W. Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 334, 1405–1408 (2011).
Gallie, D. R. The 5′-leader of tobacco mosaic virus promotes translation through enhanced recruitment of eIF4F. Nucleic Acids Res. 30, 3401–3411 (2002).
Sleat, D. E. et al. Characterisation of the 5′-leader sequence of tobacco mosaic virus RNA as a general enhancer of translation in vitro. Gene 60, 217–225 (1987).
Kim, S. H. et al. The Arabidopsis resistance-like gene SNC1 is activated by mutations in SRFR1 and contributes to resistance to the bacterial effector AvrRps4. PLoS Pathog. 6, e1001172. 10.1371/journal.ppat.1001172 (2010).
Rushton, P. J., Somssich, I. E., Ringler, P. & Shen, Q. J. WRKY transcription factors. Trends Plant Sci. 15, 247–258 (2010).
Clark, R. M. et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 317, 338–342 (2007).
Saucet, S. B. et al. Two linked pairs of Arabidopsis TNL resistance genes independently confer recognition of bacterial effector AvrRps4. Nat. Commun. 6, 6338 (2015).
Landschulz, W. H., Johnson, P. F. & McKnight, S. L. The leucine zipper: a hypothetical structure common to a new class of DNA-binding proteins. Science 240, 1759–1764 (1998).
Peart, J. R., Cook, G., Feys, B. J., Parker, J. E. & Baulcombe, D. C. An EDS1 orthologue is required for N-mediated resistance against tobacco mosaic virus. Plant J. 29, 569–579 (2002).
Liu, Y., Schiff, M., Marathe, R. & Dinesh-Kumar, S. P. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30, 415–29 (2002).
Hayward, A., Padmanabhan, M. & Dinesh-Kumar, S. P. Virus-induced gene silencing in Nicotiana benthamiana and other plant species. Methods Mol. Biol. 678, 55–63 (2011).
Sohn, K. H. et al. The nuclear immune receptor RPS4 is required for RRS1SLH1-dependent constitutive defense activation in Arabidopsis thaliana. PLoS Genet. 10, e1004655 (2014).
Zhang, X.-C. & Gassmann, W. Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol. 145, 1577–1587 (2007).
Tao, Y., Yuan, F., Leister, R. T., Ausubel, F. M. & Katagiri, F. Mutational analysis of the Arabidopsis nucleotide binding site-leucine-rich repeat resistance gene RPS2. Plant Cell 12, 2541–2554 (2000).
Chi, Y. et al. Protein-protein interactions in the regulation of WRKY transcription factors. Mol Plant. 6, 287–300 (2013).
Xu, X., Chen, C., Fan, B. & Chen, Z. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors. Plant Cell 18, 1310–1326 (2006).
Noutoshi, Y. et al. A single amino acid insertion in the WRKY domain of the Arabidopsis TIR-NBS-LRR-WRKY-type disease resistance protein SLH1 (sensitive to low humidity 1) causes activation of defense responses and hypersensitive cell death. Plant J. 43, 873–888 (2005).
Zhang, Y., Dorey, S., Swiderski, M. & Jones, J. D. Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1, SGT1 and HSP90. Plant J. 40, 213–224 (2004).
Falk, A. et al. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96, 3292–3297 (1999).
Aarts, N. et al. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signalling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 10306–10311 (1998).
Mitsuhara, I. et al. Efficient promoter cassettes for enhanced expression of foreign gene in dicotyledonous and monocotyledonous plants. Plant Cell Physiol. 37, 39–59 (1996).
Narusaka, M., Shiraishi, T., Iwabuchi, M. & Narusaka, Y. The floral inoculating protocol: a simplified Arabidopsis thaliana transformation method modified from floral dipping. Plant Biotech. 27, 349–351 (2010).
Narusaka, M., Shiraishi, T., Iwabuchi, M. & Narusaka, Y. Monitoring fungal viability and development in plants infected with Colletotrichum higginsianum by quantitative reverse transcription-polymerase chain reaction. J. Gen. Plant Pathol. 76, 1–6 (2010).
Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
Lu, R., Martin-Hernandez, M. A., Peart, J. R., Malcuit, I. & Baulcombe, D. C. Virus-induced gene silencing in plants. Methods 30, 296–303 (2003).
Dunnett, C. W. A multiple comparison procedure for comparing several treatments with a control. J. Am. Stat. Assoc. 50, 1096–1211 (1955).
Acknowledgements
This work was supported by the Science and Technology Research Promotion Program for the Agriculture, Forestry, Fisheries and Food industry awarded to Y.N., Y.T. and K.S. and by a Grant-in-Aid for Scientific Research (KAKENHI) (15K07321 to Y.N. 25450523 to M.N.). We would like to thank Yukiko Kurosaki, Yasuyo Katayama, Shoko Miyashita, Masami Miyamoto, Shoko Nieda, Aya Okada of RIBS and Atsuko Iuchi of RIKEN BRC for their excellent technical assistance, as well as Tsuyoshi Nakagawa (Shimane University) for kindly providing us with pGWB destination vectors. We also would like to thank Dr. Masaki Iwabuchi for his valuable suggestions.
Author information
Authors and Affiliations
Contributions
M.N., K.T., T.S., Y.T., K.S. and Y.N. designed research; M.N., S.I. and Y.N. performed research; and M.N. and Y.N. wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Narusaka, M., Toyoda, K., Shiraishi, T. et al. Leucine zipper motif in RRS1 is crucial for the regulation of Arabidopsis dual resistance protein complex RPS4/RRS1. Sci Rep 6, 18702 (2016). https://doi.org/10.1038/srep18702
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep18702
This article is cited by
-
DES-TOMATO: A Knowledge Exploration System Focused On Tomato Species
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.