Abstract
Study design:
We investigated microRNA (miRNA) expression after spinal cord injury (SCI) in mice.
Objectives:
The recent discovery of miRNAs suggests a novel regulatory control over gene expression during plant and animal development. MiRNAs are short noncoding RNAs that suppress the translation of target genes by binding to their mRNAs, and play a central role in gene regulation in health and disease. The purpose of this study was to examine miRNA expression after SCI.
Setting:
Department of Orthopaedic Surgery, Graduate School of Biomedical Sciences, Hiroshima University.
Methods:
We examined the expression of miRNA (miR)-223 and miR-124a in a mouse model at 6 h, 12 h, 1 day, 3 days and 7 days after SCI using quantitative PCR. The miRNA expression was confirmed by in situ hybridization.
Results:
Quantitative PCR revealed two peaks of miR-223 expression at 6 and 12 h and 3 days after SCI. MiR-124a expression decreased significantly from 1 day to 7 days after SCI. In situ hybridization demonstrated the presence of miR-223 around the injured site. However, miR-124a, which was present in the normal spinal cord, was not observed at the injured site.
Conclusion:
Our results indicate a time-dependent expression pattern of miR-223 and miR-124a in a mouse model of SCI. In this study, the time course of miRNA-223 expression may be related to inflammatory responses after SCI, and the time course of decreased miR-124a expression may reflect cell death.
Similar content being viewed by others
Introduction
Numerous studies have provided valuable information concerning the detailed cellular mechanisms and consequences of spinal cord injury (SCI). Progress continues in the development of reparative interventions to enhance recovery after experimental SCI. However, the spinal cord of adult mammals has a limited capacity for regeneration following injury.
The recent discovery of microRNAs (miRNAs) introduces a novel type of regulatory control over gene expression during plant and animal development.1 miRNAs are a family of 22-nucleotide noncoding RNAs identified in organisms ranging from nematodes to humans.1 Many miRNAs are evolutionarily conserved across phyla, and regulate gene expression by posttranscriptional gene repression. The miRNAs regulate gene expression by binding to the 3-untranslated region of the target messenger RNAs (mRNA), leading to translational repression or mRNA degradation. Several miRNAs exhibit a tissue-specific or developmental stage-specific expression pattern and have been reported to be associated with human diseases such as cancer and leukemia.2, 3 Therapeutic trials aimed at silencing miRNAs in vivo have been conducted.4
However, the expression of miRNAs in the spinal cord after SCI has not been examined and may be related to damage or renovation of the spinal cord. The purpose of the study was to examine miRNA expression patterns specific for SCI.
Materials and methods
Animals
All the experimental research protocols were reviewed and approved by the Hiroshima University ethical committee. Male C57BL/6 mice, approximately 8 weeks of age, were used (n=6 in each group). All experiments were performed according to an institutionally approved protocol in accordance with the National Institute of Health Guide for Care and Use of Laboratory Animals.
Surgical procedures
All surgical procedures were performed under 2,2,2-tribromoethanol anesthesia (125 mg kg−1 i.p., Avertin; Sigma-Aldrich, St Louis, MO, USA) using an operating microscope (Zeiss, Oberkochen, Germany). Laminectomy was carried out at the 11th thoracic vertebral level leaving the dura intact. Each animal was placed in a stereotaxic apparatus and adjustable forceps were used to atraumatically grasp the transverse process while keeping the spinal cord in a fixed position. SCI was made at the level of T11–12 by compressing the cord laterally from both sides for 10 s with a number 5 forceps (FONTAX, Lausanne, Switzerland) modified with a spacer so that a 0.5 mm space remained at maximal closure (SCI group).5 Control mice received only a laminectomy without any direct manipulation of the spinal cord (control group). Spinal cords were removed at 6 h, 12 h, 1 day, 3 days and 7 days after injury from SCI mice, and 12 h after laminectomy from control mice. Mice from both groups were anesthetized with pentobarbital sodium (100 mg kg−1 i.p) and perfused with cold saline. Spinal cords (the injured site and 5 mm on either side) were then harvested for RNA isolation as described below. Three mice each from the 12 h SCI and control groups were anesthetized and transcardially perfused with cold saline and 4% paraformaldehyde in phosphate-buffered saline (PBS; pH 7.4). The spinal cords were then removed, snap-frozen in liquid nitrogen and stored at −80 °C until histological evaluation.
RNA isolation
Total RNA was extracted from spinal cords using TRIzol regent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The concentration and quality of total RNA were measured by the UV absorbance at 260 and 280 nm (A260/280) and checked by gel electrophoresis. Equal amounts of RNA from each mouse were used for miRNA array analysis and quantitative PCR verification.
MicroRNA microarray
A miRNA microarray (Invitrogen), containing probes for the complete Sanger mirBASE 9.0, was used to screen RNA from spinal cords of mice in both the SCI and control groups killed 12 h after injury or laminectomy, respectively. Each total RNA sample was spiked with 1 μl NCode Multi-Species miRNA Microarray Controls (2 fmol μl−1), and poly(A) tailing reactions were ligated to labeled DNA polymers using the 6 × Alexa Fluor 3 (A3) or Alexa Fluor 5 (A5) Rapid Ligation Mix according to the manufacturer's protocol. The two differentially labeled reactions were combined and the volume was reduced by half in a SpeedVac Concentrator (Thermo Scientific, Waltham, MA, USA). Bovine serum albumin (50 mg ml−1) was then added to a total volume of 28.5 μl. The samples were incubated with 28.5 μl of 2 × Enhanced hybridization buffer at 65 °C for 10 min and then loaded onto NCode Multi-Species miRNA Microarrays V2. The arrays were mounted with Maui Mixer SL chambers (BioMicro Systems, Salt Lake City, UT, USA) and hybridized overnight (16–20 h) at 52 °C with constant mixing. The arrays were washed, and then scanned with a GenePix 4000B microarray scanner (Molecular Devices, Sunnyvale, CA, USA). The scanned array images were annotated and analyzed using the GenePix software and the .GAL files containing the array list (NCode_v2_mmu_gal.Gal) for the mouse samples.
Quantitative PCR verification of miRNA microarray results
Based on the results of the miRNA microarray, quantitative PCR assays were performed using a TaqMan miRNA assay kit (Applied Biosystems, Foster City, CA, USA) on RNA from the spinal cords of SCI mice killed 6 h, 12 h, 1 day, 3 days and 7 days after injury (n=6 each) and of control mice at each time point after laminectomy (n=6 each). Reverse transcription reactions of mature miRNA contained a sample of total RNA, 50 nM stem-loop RT primer, 10 × RT buffer, 100 mM each dNTPs, 50 U μl−1 MultiScribe reverse transcriptase and 20 U μl−1 RNase inhibitor. Reaction mixtures (15 μl) were incubated in a thermal cycler (MJ mini Gradient Thermal Cycler; Bio-Rad Laboratories, Hercules, CA, USA) for 30 min at 16 °C, 30 min at 42 °C and 5 min at 85 °C, and then maintained at 4 °C.
Real-time PCR was performed with a thermal cycler in 10 μl PCR mixture containing 1.33 μl RT product, 2 × TaqMan Universal PCR Master Mix, 0.2 μM TaqMan probe, 15 μM forward primer and 0.7 μM reverse primer. All reactions were incubated in triplicate in a 96-well plate at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The snoRNA-135 was used as a control to normalize differences in total RNA levels in each sample. A threshold cycle (Ct) was observed in the exponential phase of amplification, and quantification of relative expression levels was performed using standard curves for target genes and the endogenous control. Geometric means were used to calculate the ΔΔCt values and were expressed as 2−ΔΔCt. The value of each control sample was set at 1 and was used to calculate the relative expression changes in target genes.
Statistical analysis
The microarray data were analyzed according to the loop design of Kerr and Churchill.6, 7 The data of quantitative PCR assays were analyzed statistically using unpaired t-test. P-values less than 0.05 were considered statistically significant.
In situ hybridization
Spinal cords were removed at 12 h after surgery from each of three mice in the SCI and control groups, embedded in Tissue Freezing Medium (Triangle Biomedical Sciences, Durham, NC, USA) for cryostat sectioning and sectioned at 8 μm. Each section was fixed in 4% paraformaldehyde for 10 min at room temperature, washed 3 times in PBS for 3 min each and treated with 600 μg of proteinase K for 10 min at room temperature. After treatment with 0.2% glycine-PBS for 10 min, sections were refixed in 4% paraformaldehyde for 10 min, washed 3 times in PBS for 3 min each and acetylated with 0.25% acetic anhydride in 0.1 M triethanolamine hydrochloride for 10 min. After washing in PBS for 30 min, sections were prehybridized for 1 h at 65 °C with prehybridization buffer (50% formamide and 5 × saline sodium citrate (SSC)). Hybridization with locked nucleic acid DIG-labeled probes (EXIQON, VedBaek, Denmark) was carried out overnight at 65 °C in hybridization buffer (50% formamide, 5 × SSC, 5 × Denhardt's solution and 250 μg ml−1 Baker's yeast tRNA). After hybridization, sections were washed in 5 × SSC for 30 min at 65 °C, 0.2 × SSC for 2 h at 65 °C and 0.2 × SSC for 5 min at room temperature. Blocking was performed overnight at 4 °C with 4% horse serum and alkaline phosphatase-conjugated Fab anti-DIG antibody (Roche, Basel, Switzerland) in 1% sheep serum. Staining was performed using 5-bromo-4-chloro-3-indolyl-phosphate and nitroblue tetrazolium (Roche).
Results
Changes in expression of miRNAs following SCI
Using an miRNA-based array screening, we identified 10 differentially expressed miRNAs (5 decreased and 5 increased) in the SCI group compared with the control group (Table 1).
Verification and time course of the expression of miR-223 and 124a
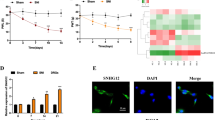
Based on the results of the miRNA microarray, we selected one increasing and one decreasing miRNA for further study. Quantitative PCR assays were performed for mmu-miR-223 and miR-124a. MiR-223 is thought to act in regulation of granulocyte production and the inflammatory response,8 and thus, miR-223 may be associated with an inflammatory reaction after SCI. MiR-124 has been reported to be expressed specifically in the central nervous system, and is considered to play a key role in the differentiation of progenitor cells to mature neurons.9 Therefore, we selected these two miRNAs for the quantitative PCR analysis. In the SCI group, the mean values (mean±s.d. (range)) for mmu-miR-223 expression were 4.58±4.45 (0.597–12.8) at 6 h, 11.7±6.07 (6.38–22.7) at 12 h, 2.26±1.32 (1.03–4.30) at 1 day, 6.92±2.93 (2.08–9.93) at 3 days and 3.70±3.40 (0.39–7.12) at 7 days after SCI. Figure 1a clearly shows that the expression of miR-223 was significantly increased at 6 h (P=0.0498), 12 h (P=0.001), and 3 days (P=0.003) after SCI compared with controls.
Summary of quantitative PCR analysis of the expression of mature miR-223 and miR-124a. (a) The expression of miR-223 was significantly increased at 6 and 12 h after spinal cord injury (SCI), only very slightly increased at 1 day, significantly increased again at 3 days and slightly increased at 7 days after SCI. (b) The expression of miR-124a was significantly decreased at 1 day after SCI and remained significantly lower than that in the control group thereafter. Error bars indicate standard deviation. *P<0.05 vs control, **P<0.01 vs control (n=6).
In the SCI group, the mean values for mmu-miR-124a expression were 1.08±0.481 (0.493–1.62) at 6 h, 0.534±0.515 (0.0483–1.20) at 12 h, 0.0792±0.0436 (0.0349–0.142) at 1 day, 0.0940±0.0857 (0.00504–0.202) at 3 days and 0.0144±0.0159 (0.000379–0.0397) at 7 days (Figure 1b). MiR-124a expression in the SCI group was significantly lower than that in the controls, 24 h after SCI (P=0.007) and thereafter (P<0.01).
In situ hybridization of miR-223 and 124a in spinal cord
MiR-223 expression was scarcely detected in the spinal cord in the control group (Figure 2a) by in situ hybridization. However, in the SCI group, miR-223 expression was present in cells distributed around the injured site (Figures 2b and c). The cells were round, with a diameter of approximately 10 μm. In contrast, miR-124a expression was identified in cells distributed along the gray matter in the control group (Figure 2d and e), and was not observed in the injured site due to a loss of tissue in the SCI group (Figure 2f).
In situ hybridization for miR-223 and miR-124a. Asterisks and arrows indicate the site of spinal cord injury and the expression of microRNAs (miRNAs), respectively. The right side of the figures is the cranial side and upper side of the figures is the dorsal side of the spinal cord. Expression of miR-223 was not observed in the control group (a). However, it was observed in cells distributed around the injured site in the spinal cord injury (SCI) group (b). The cells are round, with a diameter of approximately 10 μm (c). Expression of miR-124a was observed in cells distributed along the gray matter in the control group (d, e). However, it was not observed in the injured site due to a loss of tissue in the SCI group (f). Bars: 500 μm (a, b, d and f) and 50 μm (c and e).
Discussion
Our results showed the time course of changes in the expression of miR-223 and miR-124a, and the localization of these single nucleotide polymorphisms following SCI. Two peaks of miR-223 expression were revealed at 12 h and 3 days after SCI. In contrast, miR-124a expression was significantly decreased at 1 day through 7 days after SCI. MiR-223 was identified around the injured site by in situ hybridization. However, miR-124a, which was observed in the normal spinal cord, was not observed at the injured site in the SCI group.
MiR-223 was first identified bioinformatically, and was characterized in the hematopoietic system, where it is specifically expressed in the myeloid compartment.10 MiR-223 is thought to act as a regulator of granulocyte production and the inflammatory response.8 However, its function remains unclear. Injury to the spinal cord provokes an inflammatory reaction that results initially in further tissue damage. The mechanisms and time courses of inflammation are well documented in animal models of SCI.11 The response to trauma involves two significant waves of cellular infiltration.12 In rats, neutrophils appear at the primary lesion site at 4–6 h after injury, peak in number at 12–24 h and disappear within 5 days.13, 14 Macrophages in the injured spinal cord are derived from blood-borne monocytes and resident microglia. Blood-borne monocyte/macrophages infiltrate the lesion at 2 days after SCI in rats, achieve their highest density at 5–7 days and persist for weeks to months.11, 14 The time course of miRNA-223 expression in this study is quite similar to those of inflammation after SCI. Furthermore, the expression miR-223 is localized just around the injured site. The cells that express miR-223 were identified in the spinal cord by in situ hybridization as round and smaller than neural cells, suggesting that they are inflammatory cells. The cells were found at 12 h after injury, and, therefore, they may be neutrophils, particularly granulocytes. The increasing miR-223 expression at 3 days after SCI also may relate to the appearance of macrophages in the injured site, although we did not fully characterize the cells in this study.
MiR-124 is expressed specifically in muscle and the central nervous system, and is considered to play a key role in the differentiation of progenitor cells to mature neurons.15 In this study, miR-124a expression was identified in cells distributed along the gray matter in the control and SCI group, by in situ hybridization. Astrocytes exist in both gray and white matter and oligodendrocyte exist only in the white matter. Cells that express miR-124a were identified in the gray matter only, suggesting that they are neurons. Our data show a significant decrease in miR-124a expression at 12 h after SCI that persists at least until day 7. Cell death after SCI results not only from the physical trauma to the tissue, but also from secondary injury that expands the damage to levels of the cord rostral and caudal to the actual impact site. The inflammatory response is likely to be an important factor in the development of secondary damage.14 Secondary cell death has been reported to be virtually complete at 12 h.16 The time course of decreased miR-124a expression seen in this study may reflect cell death after SCI.
In this study, the miRNA-based array screening also shows increased miR-1, miR-133a, miR-133b and miR-451 expressions, and decreased miR-129-3p, miR-342, miR-495 and miR-541 expressions. MiR-1 and miR-133 have distinct roles in modulating skeletal and cardiac muscle proliferation and differentiation.17 Furthermore, Ivey et al.18 demonstrated that miR-1 and miR-133 may curtail the differentiation of pluripotent cells into mature neurons, even as cells are pushed toward that lineage by timed administration of retinoic acid. Thus, these miRNAs may relate to neural responses after SCI. Although the function of other miRNAs could not be identified sufficiently, miR-451, miR-129 and miR-342 are reported to relate to differentiation of blood cells.19, 20 Therefore, these miRNAs variations may appear as responses after bleeding in the injured site.
Recently, several in vivo studies provided evidence suggesting the potential therapeutic usefulness of silencing miRNA.4 Therefore, further functional analyses to define the precise role of miRNAs in the pathogenesis of SCI may provide novel attractive therapeutic tools for SCI that regulate miRNA expression.
References
Ambros V . The functions of animal microRNAs. Nature 2004; 431: 350–355.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435: 834–838.
Marcucci G, Radmacher MD, Maharry K, Mrozek K, Ruppert AS, Paschka P et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med 2008; 358: 1919–1928.
Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005; 438: 685–689.
Faulkner JR, Herrmann JE, Woo MJ, Tansey KE, Doan NB, Sofroniew MV . Reactive astrocytes protect tissue and preserve function after spinal cord injury. J Neurosci 2004; 24: 2143–2155.
Altman NS, Hua J . Extending the loop design for two-channel microarray experiments. Genet Res 2006; 88: 153–163.
Kerr MK, Martin M, Churchill GA . Analysis of variance for gene expression microarray data. J Comput Biol 2000; 7: 819–837.
Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 2008; 451: 1125–1129.
Kosik KS . The neuronal microRNA system. Nat Rev Neurosci 2006; 7: 911–920.
Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP . Vertebrate microRNA genes. Science 2003; 299: 1540.
Popovich PG, Wei P, Stokes BT . Cellular inflammatory response after spinal cord injury in Sprague-Dawley and Lewis rats. J Comp Neurol 1997; 377: 443–464.
Blight AR . Macrophages and inflammatory damage in spinal cord injury. J Neurotrauma 1992; 9 (Suppl 1): S83–S91.
Taoka Y, Okajima K, Uchiba M, Murakami K, Kushimoto S, Johno M et al. Role of neutrophils in spinal cord injury in the rat. Neuroscience 1997; 79: 1177–1182.
Carlson SL, Parrish ME, Springer JE, Doty K, Dossett L . Acute inflammatory response in spinal cord following impact injury. Exp Neurol 1998; 151: 77–88.
Makeyev EV, Zhang J, Carrasco MA, Maniatis T . The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell 2007; 27: 435–448.
Dusart I, Schwab ME . Secondary cell death and the inflammatory reaction after dorsal hemisection of the rat spinal cord. Eur J Neurosci 1994; 6: 712–724.
Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 2006; 38: 228–233.
Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell 2008; 2: 219–229.
Rathjen T, Nicol C, McConkey G, Dalmay T . Analysis of short RNAs in the malaria parasite and its red blood cell host. FEBS Lett 2006; 580: 5185–5188.
Liao R, Sun J, Zhang L, Lou G, Chen M, Zhou D et al. MicroRNAs play a role in the development of human hematopoietic stem cells. J Cell Biochem 2008; 104: 805–817.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nakanishi, K., Nakasa, T., Tanaka, N. et al. Responses of microRNAs 124a and 223 following spinal cord injury in mice. Spinal Cord 48, 192–196 (2010). https://doi.org/10.1038/sc.2009.89
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sc.2009.89
Keywords
This article is cited by
-
Spinal Cord Injury: From MicroRNAs to Exosomal MicroRNAs
Molecular Neurobiology (2024)
-
Correlation between miRNA-124, miRNA-544a, and TNF-α levels in acute spinal cord injury
Spinal Cord (2022)
-
miR-495 reduces neuronal cell apoptosis and relieves acute spinal cord injury through inhibiting PRDM5
Journal of Molecular Histology (2021)
-
The inhibition by human MSCs-derived miRNA-124a overexpression exosomes in the proliferation and migration of rheumatoid arthritis-related fibroblast-like synoviocyte cell
BMC Musculoskeletal Disorders (2020)
-
Nociceptive related microRNAs and their role in rheumatoid arthritis
Molecular Biology Reports (2020)