Abstract
The oriental armyworm, Mythimna separata, is an extremely destructive polyphagous pest with a broad host range that seriously threatens the safety of agricultural production. Here, a high-quality chromosome-level genome was assembled using Illumina, PacBio HiFi long sequencing, and Hi-C scaffolding technologies. The genome size was 706.30 Mb with a contig N50 of 22.08 Mb, and 99.2% of the assembled sequences were anchored to 31 chromosomes. In addition, 20,375 protein-coding genes and 258.68 Mb transposable elements were identified. The chromosome-level genome assembly of M. separata provides a significant genetic resource for future studies of this insect and contributes to the development of management strategies.
Similar content being viewed by others
Background & Summary
The oriental armyworm, Mythimna separata (Lepidoptera, Noctuidae), is a notorious polyphagous pest that is widely distributed in Asia, Australia, New Zealand, and several Pacific islands1,2,3 (Fig. 1a). This pest has a wide host range and poses a serious threat to the production of crops, particularly rice, maize, and wheat4 (Fig. 1b). The outbreak of M. separata in China from 2012 to 2013 threatened 1743.7 million hectares of farmland5, and this threat has continued in recent years6,7,8. This situation also occurs in other countries and regions where M. separata infestations are present9. In recent years, with the changes in global climate, crop planting structure, variety distribution, and cultivation system, M. separata has shown new characteristics in adaptability, breakout, and damage10,11. Due to its gregariousness, migration capability, polyphagy, and gluttony, M. separata was included in the list of first-class crop diseases and insect pests by the Chinese Ministry of Agriculture and Rural Affairs in 2020.
Previous studies have shown that polyphagous insects respond to toxic secondary metabolites produced by different host plants by inducing changes in the expression of genes related to detoxifying enzymes. Such changes may enhance the ability of polyphagous insects to adapt to host plants and develop resistance against pesticides12. However, the scarcity of genomic resources prevents the above hypothesis from being verified in M. separata. Although several M. separata genome assemblies were published in 2022 and 202313,14,15,16, there are significant differences in the assembly method and quality of these genome assemblies. Hence, a high-quality chromosomal level genome is necessary to offer genetic resources and delve into the molecular mechanism of detoxification and host adaptation of M. separata, which will aid in providing theoretical support for optimizing management strategies for M. separata.
In the present study, we assembled a high-quality chromosome-level genome of M. separata by using a combination of Illumina short reads, PacBio high fidelity (HiFi) reads, and high-throughput chromosome conformation capture (Hi-C) data (Table 1). The genome assembly consisted of 172 contigs with a total length of 706.30 Mb, of which the contig N50 was 22.08 Mb. In addition, 99.2% of the draft assembly (700.63 Mb) was anchored to 31 chromosomes with a scaffold N50 of 23.00 Mb. We also identified 258.68 Mb of tandem repeats, accounting for 36.63% of the genome assembly. A total of 20,375 protein-coding genes were obtained, of which 98.53% were annotated. The results of phylogenetic analysis revealed that M. separata was diverged from Helicoverpa armigera approximately 25.91 Mya. Furthermore, 594 expanded gene families and 1329 contracted gene families were identified in M. separata genome. The high-quality chromosome-level genome assembly of M. separata will provide a genetic basis for further research on this polyphagous pest.
Methods
Sample collection and genome sequencing
M. separata was collected from maize fields in Anyang City, Henan Province, China, and was subsequently reared in climate incubators at a temperature of 26 ± 1 °C with a relative humidity of 70% and a photoperiod of 14 h L:10 h D17. Genomic DNA was extracted from a single surface-sterilized male pupa using the QIAamp DNA Mini Kit (QIAGEN) for both Illumina and PacBio HiFi sequencing to prevent contamination from other individuals and microorganisms. For Hi-C sequencing, genomic DNA was extracted from a single male adult. Total RNA was extracted from adults using the TRIzol kit for transcriptome sequencing. The purity and integrity of genomic DNA and RNA were validated by the NanoDrop 2000C spectrophotometer (Thermo, Wilmington, DE, USA) and agarose gel electrophoresis (1.5%).
The paired-end libraries with a 350 bp inserted fragment were constructed and sequenced on the Illumina NovaSeq6000 platform following the manufacturer’s instruction. After removing adapter sequences and low-quality reads with HTQC (v1.92.310) software18, a total of 58.72 Gb clean reads were obtained for subsequent analyses. For PacBio HiFi sequencing, genomic DNA was sheared into ~15 Kb fragments using g-Tubes (Covaris, Woburn, MA, USA) and purified using 0.45 × AMPure PB beads (Beckman Coulter, Brea, CA, USA) for constructing SMRT bell libraries. Size selection was performed using the Sage ELF system (Sage Science, Beverly, MA) to collect SMRT bell libraries of 15–18 Kb. After annealing primers and binding Sequel II DNA polymerase to SMRT bell templates, sequencing was performed using 8 M SMRT cells on the Sequel II System (Biomarker Technologies Co., LTD, Beijing, China). A total of 986.03 Gb subreads were obtained and utilized to generate PacBio HiFi reads via the circular consensus sequencing (CCS) mode. Finally, a total of 70.62 Gb of CCS reads were produced, with an average read length of 16.67 kb, resulting in 99.98X coverages of the M. separata genome. The Hi-C library was constructed following the standard library preparation protocol19 and sequenced on the Illumina NovaSeq6000 platform, and 76.08 Gb of 150-bp paired-end clean reads were obtained.
Genome survey and assembly
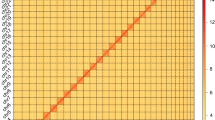
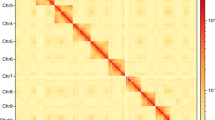
Genome survey was essential to estimate the main characteristics, including genome size, repetitive sequence content and heterozygosity. The k-mer (K = 19) frequencies were constructed based on Illumina clean short-reads using Jellyfish (v2.2.10)20 and used to perform genome survey by GenomeScope (v2.0)21. The estimated genome scale of M. separata was 662.64 Mb, with a repetitive content of 39.00% and a heterozygosity of 0.76% (Fig. 2a). Subsequently, CCS reads were submitted to Hifiasm (v0.15.1)22 and assembled with default parameters. After filtering haplotypic duplicates using purge_dups23 with parameters of ‘−2 -T cutoffs -c PB.base.cov’, the M. separata genome assembly was generated. The assembly consisted of 172 contigs with a total length of 706.30 Mb and a contig N50 of 22.08 Mb. The clean Hi-C reads were aligned to the draft genome assembly using BWA (0.7.10)24 with default parameters. The uniquely aligned read pairs were further processed using HiC-Pro (v2.10.0)25 to assess and eliminate the invalid read pairs, including dangling-end, re-ligation, self-cycle, and dumped pairs. A total of 88,824,108 valid interaction pairs for scaffold correction were used to cluster, order, and orient contigs onto chromosomes using LACHESIS (v2e27abb)26 with default parameters. Finally, 147 scaffolds were anchored to 31 chromosomes with a scaffold N50 of 23.00 Mb, covering a span of 700.63 Mb and representing 99.2% of the draft genome assembly (Fig. 2b,c, Table 2). In addition, the mitochondrial genome of M. separata was assembled through mitoZ27 and NOVOplasty28, and subsequently annotated using MITOS29 and GeSeq30 (Fig. 3a, Table 3).
Genome assembly of M. separata. (a) Genome scope profiles of 19-mer analysis. (b) Circle genome landscape of M. separata. Circle I represents chromosomes, while circles II-IV indicate repeat density, gene density, and GC content of each respective chromosome. (c) Hi-C interactive heatmap of M. separata. Color indicates the intensity of the interaction signal. The darker the color, the higher the intensity.
Genomic repeat annotation
Repeat sequences mainly include tandem repeats and interspersed repeats, with the latter mainly being transposable elements (TE). The repeat sequences of TE were annotated using a combination of homology-based and de novo approaches. We initially customized a de novo repeat library using RepeatModeler31 and LTR_retriever (v2.8)32 based on the assembly sequences with default parameters. The predicted repeats were subsequently classified using PASTEClassifier (v1.0)33, and the results were combined with databases of Repbase34, REXdb (v3.0)35, and Dfam (v3.2)36 to construct a species-specific TE library without redundancy. TE sequences were identified by homology search against the library using RepeatMasker (v4.10)37. A total of 258.68 Mb TE sequences were obtained, accounting for 36.63% genome assembly. In addition, 23.64 Mb (3.35%) tandem repeats were identified using MISA (v2.1)38 and NCRF39 (Table 4).
Gene prediction and functional annotation
Three approaches, including de novo prediction, homolog-based and transcriptome-based methods, were combined to perform gene prediction after eliminating the interference of repeat sequences in the M. separata genome. The de novo gene models were predicted using two ab initio gene-prediction software tools of Augustus (v2.4)40 and SNAP41 with default parameters. Homology-based gene prediction was conducted using GeMoMa (v1.7)42 against the protein sequences of lepidopteran insects B. mori, H. armigera, S. frugiparda, and S. litura downloaded from GenBank. For transcriptome-based gene prediction, the RNA-seq reads were assembled into unigenes using Trinity (v2.11)43, and resulting unigenes were then used to identify protein-coding genes via PASA (v2.0.2)44. Finally, gene models obtained from these three methods were integrated into a unified gene set using EVidenceModeler (v1.1.1)45 with default parameters. As a result, 20,375 protein-coding genes were identified from M. separata genome (Fig. 3b).
In order to perform functional annotation of the protein-coding genes, we aligned predicted genes against databases including NR, GO, KEGG, EggNOG, KOG, TrEMBL, InterPro and Swiss-Prot using BLAST (v2.2.31) with a threshold of 1e−5. Finally, 98.53% (20075/20375) of protein-coding genes were annotated (Table 5). The detoxification-related genes cytochrome P450 (P450), ATP-binding cassette (ABC), Carboxyl/cholinesterase (CCE), UDP-glycosyltransferases (UGTs), and glutathione-S-transferase (GST), as well as the chemosensory-related genes of ionotropic receptors (IRs), chemosensory proteins (CSPs), and odorant binding proteins (OBPs), were further annotated using BLASTP (E < 10−5). To annotate genes associated with gustatory receptors (GRs) and odorant receptors (ORs), we identified candidate loci through TBLASTN with E-values < 10−5 and predicted gene structures using GeneWise (v2.2.0)46 (Fig. 4).
Divergence time and distribution of detoxification and chemosensory genes in M. spearata and other eight lepidopteran insects. The branch node values indicate the inferred divergence time between species. The numbers in the right cells indicate the scale of the corresponding gene family in each species. The darker the background color of cells, the more the genes encoded in the corresponding species.
Phylogenetic analysis
The protein sequences of seventeen insects, including eight Lepidoptera insects and nine others associated with Diptera, Coleoptera, Hymenoptera, Hemiptera, and Odonata, were downloaded from NCBI for phylogenetic analysis (Table 6). The orthologous gene families were detected using OrthoFinder (v2.4.0)47 and annotated based on the PANTHER48 database. The single-copy orthologous genes were aligned using MAFFT (v7.205)49, and ambiguously aligned regions were removed by applying Gblocks (v0.91b)50 with default parameters. The phylogenetic trees were constructed by IQ-TREE (v1.6.10)51 with 1000 bootstrap replicates and the best model of LG + F + I + G4. The divergence time between different species was estimated using MCMCtree (PAML52 package) based on the fossil records acquired from TimeTree database (http://www.timetree.org/). Furthermore, the results obtained from phylogenetic trees, which included divergence time, were employed to identify the expansion and contraction of gene families using CAFE (v5.0)53 with a p-value threshold ≤ 0.05.
Genome synteny analysis
In order to perform genome synteny analysis of M. separata with Spodoptera frugiperda, the similar gene pairs were identified using Diamond (v0.9.29)54 with default parameters. All genes in synteny blocks were obtained by MCScanX55, and synteny blocks were then visualized across chromosomes using CIRCOS (v 0.69–9)56. Only one fission event was identified between M. separata and Spodoptera frugiperda, which suggested a high degree of concordance between them (Fig. 5a).
Data Records
The raw data of Illumina sequencing, PacBio HiFi sequencing and Hi-C sequencing of the Mythimna separata genome was deposited at the NCBI SRA database with the accession number of SRP43304057. The final assembled Mythimna separata genome has been submitted to NCBI under accession number GCA_030763345.158. The annotation files of the Mythimna separata genome have been deposited at figshare59.
Technical Validation
Evaluation of the genome assembly
The integrity and accuracy of genome assembly were verified from three aspects: Firstly, the clean reads acquired from Illumina sequencing were aligned against the genome assembly using BWA24. The results revealed that 99.26% of Illumina reads were aligned to the genome assembly. Secondly, the Core Eukaryotic Genes Mapping Approach (CEGMA) database contained 458 conserved core eukaryotic genes, of which 431 (94.10%) were identified in M. separata genome. Finally, the completeness of the genome assembly was evaluated using BUSCO (v4)20 with parameters of ‘-m prot -f -l eukaryota_odb9’, and 98.74% of the conserved core BUSCOs were identified in the genome of M. separata. These results showed that we obtained the high-quality M. separata genome assembly. Meanwhile, the contig N50 in our assembly was 22.08 Mb, which was significantly higher than the 7.31 Mb in recent assembly version of M. separata13. The scaffold N50 in our assembly was improved to 23.00 Mb, which was slightly higher than the 22.68 Mb in other recent assembly version of M. separata15 (Table 7).
To assess the quality of chromosome assembly, the assembly was sheared into 100 kb bins, and the intensity of the interaction pairs was used to plot heatmaps. The Hi-C heatmap showed that the intensity of interaction along the diagonals was obviously higher than that at non-diagonal positions in 31 distinct chromosomes.
Evaluation of gene prediction
The BUSCO analysis was also used to assess the results of gene prediction. The 98.74% (942/954) of the BUSCOs were identified from the predicted gene set of our genome, which was slightly higher than the 98% and 98.2% in other recent M. separata assembly version13,15. Meanwhile, 83.84% of the RNA-seq data were aligned to the predicted exons (Fig. 5b). These results confirmed the completeness and accuracy of gene prediction across M. separata genome. In addition, 20,375 protein-coding genes were identified in our genome assembly, which was significantly more than the 17549 protein-coding genes in the best available recent reference genome assembly version of M. separata15. We further compared the set of protein-coding genes in the two genome assemblies using local BLASTN with E-values < 10−5. A total of 16,398 protein-coding genes were identified in both two genome assemblies, and 2,828 protein-coding genes were identified only in our genome assembly.
Comparative genomic analysis
A total of 27,002 orthologous gene families were identified from the 18 insect species, of which 565 single-copy orthologous gene families were used for phylogenetic analysis (Table S1). The results of phylogenetic analysis indicated that Lepidoptera insects speciated from their common ancestor later than Diptera, Coleoptera, Hymenoptera and Hemiptera (Fig. 6). M. separata and H. armigera were found to cluster into a single clade within Lepidoptera and to diverge at approximately 25.29 (20.57–30.31) million years ago (mya). In addition, M. separata and S. frugiperda were estimated to diverge at approximately 33.82 (29.02–36.67) Mya ago. Meanwhile, GO enrichment analysis revealed that the 594 expanded gene families in M. separata genome mainly involved “DNA integration” (GO:0015074), “nucleosome” (GO: 0000786), and “RNA-directed DNA polymerase activity” (GO:0003964), while the 1329 contracted gene families mainly involved “regulation of signal transduction” (GO:0009966), “membrane” (GO:0016020), and “serine-type endopeptidase activity” (GO:0004252) (Figs. 7, 8, Tables S2, S3).
Phylogenetic tree of M. spearata together with 17 other insects. The maximum likelihood phylogenomic tree was calculated based on 565 single-copy genes. The numbers of expanded gene families (green) and contracted gene families (red) are displayed to the right of each species branch. The coloured histogram indicates that genes of each species were categorized into five groups: 1:1:1 (single copy orthologous genes in common gene families); N: N: N (multiple copy orthologous genes in common gene common gene families); Specific (genes from unique gene families from each species); Other (genes that do not belong to any of the above ortholog categories); Unclustered (genes which are not clustered into any family).
Code availability
All bioinformatic tools and softwares for data analysis in this study were used according to the manuals, and the version and code/parameters of software have been introduced in Methods section. No custom code was used.
References
Sharma, H. C. & Davies, J. C. The oriental armyworm, Mythimna separata (Wlk.). Distribution, biology and control: a literature review. Miscellaneous Reports - Centre for Overseas Pest Research (UK). 59, 24 (1983).
Jiang, X. F., Zhang, L., Cheng, Y. X. & Luo, L. Z. Current status and trends in research on the oriental armyworm, Mythimna separata (Walker) in China. Chinese Journal of Applied Entomology. 51, 881–889 (2014).
Sharma, H. C., Sullivan, D. J. & Bhatnagar, V. S. Population dynamics and natural mortality factors of the Oriental armyworm, Mythimna separata (Lepidoptera: Noctuidae), in South-Central India. Crop Protection. 21, 721–732 (2002).
Wang, G. P., Zhang, Q. W., Ye, Z. H. & Luo, L. Z. The role of nectar plants in severe outbreaks of armyworm Mythimna separata (Lepidoptera: Noctuidae) in China. Bulletin of Entomological Research. 96, 445–455 (2006).
Ertao, L. I. et al. Control efficacy of the combined application of the entomopathogenic nematode and Bt against the oriental armyworm, Mythimna separata (Walker). Plant Protection. 45, 295–302 (2019).
Jie Liu, Y. J., Huang, C., Wu, Q., Zhang, T. & Zeng, J. Forecast on the occurrence trend of major diseases and insect pests of grain crops in China in 2021. China Plant Protection. 042, 37–39 (2021).
Jie Liu, T. L., Jiang, Y., Zeng, J. & Wang, Y. Occurrence characteristics of main maize diseases and insect pests in china in 2020. China Plant Protection. 041, 30–35 (2021).
Jie Liu, J. Z., Yang, Q., Bian, Y., Zhang, Y. & Huang, C. Forecast of occurrence trend of major crop diseases and insect pests in 2023. China Plant Protection. 43, 32–35 (2023).
Sharma, H. C. & Davies, J. C. The oriental armyworm, Mythimna separata (Wlk.). Distribution, biology and control: a literature review. Miscellaneous Reports - Centre for Overseas Pest Research (UK) (1983).
Liu, Y. et al. Chitin deacetylase: A potential target for Mythimna separata (Walker) control. Arch Insect Biochem Physiol. 104, e21666 (2020).
Jiang, X., Luo, L., Zhang, L., Sappington, T. W. & Hu, Y. Regulation of migration in Mythimna separata (Walker) in China: a review integrating environmental, physiological, hormonal, genetic, and molecular factors. Environ Entomol. 40, 516–533 (2011).
de la Paz Celorio-Mancera, M. et al. Mechanisms of macroevolution: polyphagous plasticity in butterfly larvae revealed by RNA-Seq. Mol Ecol. 22, 4884–4895 (2013).
Tong, D. et al. The oriental armyworm genome yields insights into the long-distance migration of noctuid moths. Cell Rep. 41, 111843 (2022).
Yokoi, K., Furukawa, S., Zhou, R., Jouraku, A. & Bono, H. Reference Genome Sequences of the Oriental Armyworm, Mythimna separata (Lepidoptera: Noctuidae). Insects. 13, 1172 (2022).
Zhao, H. et al. Chromosome-level genomes of two armyworms, Mythimna separata and Mythimna loreyi, provide insights into the biosynthesis and reception of sex pheromones. Mol Ecol Resour. 23, 1423–1441 (2023).
NCBI Assembly https://www.ncbi.nlm.nih.gov/assembly/GCA_020882275.1 (2021).
Wang, Y.-r. et al. Unveiling sugarcane defense response to Mythimna separata herbivory by a combination of transcriptome and metabolic analyses. Pest Management Science. 77, 4799–4809 (2021).
Yang, X. et al. HTQC: a fast quality control toolkit for Illumina sequencing data. BMC bioinformatics. 14, 33 (2013).
Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 159, 1665–1680 (2014).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31, 3210–3212 (2015).
Vurture, G. W. et al. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics. 33, 2202–2204 (2017).
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nature Methods. 18, 170–175 (2021).
Guan, D. et al. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics. 36, 2896–2898 (2020).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25, 1754–1760 (2009).
Servant, N. et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 259 (2015).
Burton, J. N. et al. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nature Biotechnology. 31, 1119–1125 (2013).
Meng, G., Li, Y., Yang, C. & Liu, S. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47, e63 (2019).
Dierckxsens, N., Mardulyn, P. & Smits, G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45, e18 (2017).
Bernt, M. et al. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69, 313–319 (2013).
Tillich, M. et al. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, W6–w11 (2017).
Flynn, J. M. et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci USA 117, 9451–9457 (2020).
Ou, S. & Jiang, N. LTR_retriever: A Highly Accurate and Sensitive Program for Identification of Long Terminal Repeat Retrotransposons. Plant Physiol. 176, 1410–1422 (2018).
Hoede, C. et al. PASTEC: an automatic transposable element classification tool. PLoS One. 9, e91929 (2014).
Bao, W., Kojima, K. K. & Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 6, 11 (2015).
Neumann, P., Novák, P., Hoštáková, N. & Macas, J. Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mob DNA. 10, 1 (2019).
Wheeler, T. J. et al. Dfam: a database of repetitive DNA based on profile hidden Markov models. Nucleic Acids Res. 41, D70–82 (2013).
Tarailo-Graovac, M. & Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics. Chapter 4, Unit 4.10 (2009).
Beier, S., Thiel, T., Münch, T., Scholz, U. & Mascher, M. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33, 2583–2585 (2017).
Harris, R. S., Cechova, M. & Makova, K. D. Noise-cancelling repeat finder: uncovering tandem repeats in error-prone long-read sequencing data. Bioinformatics. 35, 4809–4811 (2019).
Stanke, M., Diekhans, M., Baertsch, R. & Haussler, D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 24, 637–644 (2008).
Korf, I. Gene finding in novel genomes. BMC Bioinformatics. 5, 59 (2004).
Keilwagen, J. et al. Using intron position conservation for homology-based gene prediction. Nucleic Acids Res. 44, e89 (2016).
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z. & Amit, I. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nature Biotechnology. 29, 644 (2013).
Haas, B. J. et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31, 5654–5666 (2003).
Haas, B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 9, R7 (2008).
Birney, E., Clamp, M. & Durbin, R. GeneWise and Genomewise. Genome Res. 14, 988–995 (2004).
Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019).
Mi, H., Muruganujan, A., Ebert, D., Huang, X. & Thomas, P. D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–d426 (2019).
Katoh, K., Asimenos, G. & Toh, H. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol. 537, 39–64 (2009).
Talavera, G. & Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56, 564–577 (2007).
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32, 268–274 (2015).
Yang, Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 13, 555–556 (1997).
Mendes, F. K., Vanderpool, D., Fulton, B. & Hahn, M. W. CAFE 5 models variation in evolutionary rates among gene families. Bioinformatics. 36, 5516–5518 (2020).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12, 59–60 (2015).
Wang, Y. et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49 (2012).
Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009).
NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP433040 (2023).
NCBI Assembly https://identifiers.org/ncbi/insdc.gca:GCA_030763345.1 (2023).
Xu, C. The annotation data of Mythimna separata genome. figshare https://doi.org/10.6084/m9.figshare.23658951.v1 (2023).
Acknowledgements
This work is supported by National Key R&D Program of China (2022YFD1400300) and Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences.
Author information
Authors and Affiliations
Contributions
J.J., J.C., J.L. and X.G. conceived the project; C.X., X.Z., L.W., K.Z. and D.L. performed the experiments; C.X., N.H. and H.X. performed the bioinformatic analyses; C.X., R.C. and L.N. evaluated the results; C.X. and J.J. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xu, C., Ji, J., Zhu, X. et al. Chromosome level genome assembly of oriental armyworm Mythimna separata. Sci Data 10, 597 (2023). https://doi.org/10.1038/s41597-023-02506-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-023-02506-3
This article is cited by
-
Beyond NGS data sharing for plant ecological resilience and improvement of agronomic traits
Scientific Data (2024)
-
Chromosome-level genome assembly of the northern Pacific seastar Asterias amurensis
Scientific Data (2023)