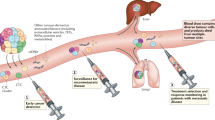
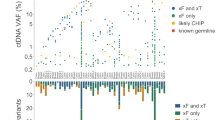
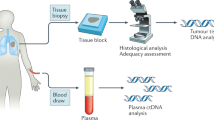
Abstract
Anticancer drug development is a costly and protracted activity, and failure at late phases of clinical testing is common. We have previously proposed the Pharmacological Audit Trail (PhAT) intended to improve the efficiency of drug development, with a focus on the use of tumour tissue-based biomarkers. Blood-based ‘liquid biopsy’ approaches, such as targeted or whole-genome sequencing studies of plasma circulating cell-free tumour DNA (ctDNA) and circulating tumour cells (CTCs), are of increasing relevance to this drug development paradigm. Liquid biopsy assays can provide quantitative and qualitative data on prognostic, predictive, pharmacodynamic and clinical response biomarkers, and can also enable the characterization of disease evolution and resistance mechanisms. In this Perspective, we examine the promise of integrating liquid biopsy analyses into the PhAT, focusing on the current evidence, advances, limitations and challenges. We emphasize the continued importance of analytical validation and clinical qualification of circulating tumour biomarkers through prospective clinical trials.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Sharpe, E., Hoey, R., Yap, C. & Workman, P. From patent to patient: analysing access to innovative cancer drugs. Drug Discov. Today 25, 1561–1568 (2020).
Biotechnology Innovation Organization, Biomedtracker & Amplion. Clinical Development Success Rates 2006-2015 bio.org https://www.bio.org/sites/default/files/legacy/bioorg/docs/Clinical%20Development%20Success%20Rates%202006-2015%20-%20BIO,%20Biomedtracker,%20Amplion%202016.pdf (2016).
Workman, P., Draetta, G. F., Schellens, J. H. M. & Bernards, R. How much longer will we put up with 100,000 cancer drugs? Cell 168, 579–583 (2017).
Workman, P. How much gets there and what does it do?: The need for better pharmacokinetic and pharmacodynamic endpoints in contemporary drug discovery and development. Curr. Pharm. Des. 9, 891–902 (2005).
Yap, T. A., Sandhu, S. K., Workman, P. & de Bono, J. S. Envisioning the future of early anticancer drug development. Nat. Rev. Cancer 10, 514 (2010).
Workman, P. Auditing the pharmacological accounts for Hsp90 molecular chaperone inhibitors: unfolding the relationship between pharmacokinetics and pharmacodynamics. Mol. Cancer Ther. 2, 131–138 (2003).
Banerji, U. et al. Phase I pharmacokinetic and pharmacodynamic study of 17-allylamino, 17-demethoxygeldanamycin in patients with advanced malignancies. J. Clin. Oncol. 23, 4152–4161 (2005).
Attard, G. et al. Phase I clinical trial of a selective inhibitor of CYP17, abiraterone acetate, confirms that castration-resistant prostate cancer commonly remains hormone driven. J. Clin. Oncol. 26, 4563–4571 (2008).
Attard, G. et al. Selective inhibition of CYP17 with abiraterone acetate is highly active in the treatment of castration-resistant prostate cancer. J. Clin. Oncol. 27, 3742–3748 (2009).
Fong, P. C. et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 361, 123–134 (2009).
Sarker, D. et al. First-in-human phase I study of pictilisib (GDC-0941), a potent pan-class I phosphatidylinositol-3-kinase (PI3K) inhibitor, in patients with advanced solid tumors. Clin. Cancer Res. 21, 77–86 (2015).
Yap, T. A. et al. First-in-class phase I trial of a selective Akt inhibitor, MK2206 (MK), evaluating alternate day (QOD) and once weekly (QW) doses in advanced cancer patients (pts) with evidence of target modulation and antitumor activity [abstract]. J. Clin. Oncol. 28 (Suppl. 15), 3009 (2010).
Raynaud, F. I. et al. Biological properties of potent inhibitors of class I phosphatidylinositide 3-kinases: from PI-103 through PI-540, PI-620 to the oral agent GDC-0941. Mol. Cancer Ther. 8, 1725–1738 (2009).
Carden, C. P., Banerji, U., Kaye, S. B., Workman, P. & de Bono, J. S. From darkness to light with biomarkers in early clinical trials of cancer drugs. Clin. Pharmacol. Ther. 85, 131–133 (2009).
Carden, C. P. et al. Can molecular biomarker-based patient selection in phase I trials accelerate anticancer drug development? Drug Discov. Today 15, 88–97 (2010).
de Bono, J. S. et al. Randomized phase II study evaluating Akt blockade with ipatasertib, in combination with abiraterone, in patients with metastatic prostate cancer with and without PTEN loss. Clin. Cancer Res. 25, 928–936 (2019).
Ferraldeschi, R. et al. PTEN protein loss and clinical outcome from castration-resistant prostate cancer treated with abiraterone acetate. Eur. Urol. 67, 795–802 (2015).
Biondo, A. et al. Research related tumour biopsies in early-phase trials with simultaneous molecular characterisation – a single unit experience. Cancer Treat. Res. Commun. 27, 100309 (2021).
Perez-Lopez, R. et al. Diffusion-weighted imaging as a treatment response biomarker for evaluating bone metastases in prostate cancer: a pilot study. Radiology 283, 168–177 (2017).
Goodall, J. et al. Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov. 7, 1006–1017 (2017).
Attard, G. et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 69, 2912–2918 (2009).
Banerji, U. & Workman, P. Critical parameters in targeted drug development: the pharmacological audit trail. Semin. Oncol. 43, 436–445 (2016).
Rossanese, O. et al. The pharmacological audit trail (PhAT): Use of tumor models to address critical issues in the preclinical development of targeted anticancer drugs. Drug Discov. Today Dis. Model. 21, 23–32 (2016).
Takimoto, C. H. Pharmacokinetics and pharmacodynamic biomarkers in early oncology drug development. Eur. J. Cancer 45, 436–438 (2009).
Seyhan, A. A. Biomarkers in drug discovery and development. Eur. Pharm. Rev. 5, 19–25 (2010).
Hait, W. N. Forty years of translational cancer research. Cancer Discov. 1, 383–390 (2011).
Garralda, E., Dienstmann, R. & Tabernero, J. Pharmacokinetic/pharmacodynamic modeling for drug development in oncology. Am. Soc. Clin. Oncol. Educ. Book 37, 210–215 (2017).
Takimoto, C. & Wick, M. in Principles of Clinical Pharmacology Ch. 31 (eds Arthur, J. & Huang, S.-M.) 517–529 (Academic, 2012).
Eisenhauer, E. A., Twelves, C. & Buyse, M. Phase I Cancer Clinical Trials: A Practical Guide (Oxford Univ. Press, 2015).
Hait, W. in Cancer Medicine Ch. 27 (ed. Hong, W.) (People’s Medical Publishing House, 2010).
Sandhu, S. K. et al. The poly(ADP-ribose) polymerase inhibitor niraparib (MK4827) in BRCA mutation carriers and patients with sporadic cancer: a phase 1 dose-escalation trial. Lancet. Oncol. 14, 882–892 (2013).
de Bono, J. et al. Phase I, dose-escalation, two-part trial of the PARP inhibitor talazoparib in patients with advanced germline BRCA1/2 mutations and selected sporadic cancers. Cancer Discov. 7, 620–629 (2017).
Yap, T. A. et al. Phase I trial of first-in-class ATR inhibitor M6620 (VX-970) as monotherapy or in combination with carboplatin in patients with advanced solid tumors. J. Clin. Oncol. 38, 3195–3204 (2020).
Slovin, S. F. et al. Pharmacodynamic and clinical results from a phase 1/2 study of the HSP90 inhibitor onalespib in combination with abiraterone acetate in prostate cancer. Clin. Cancer Res. 25, 4624–4633 (2019).
Levit, L. A. et al. Ethical framework for including research biopsies in oncology clinical trials: American Society of Clinical Oncology research statement. J. Clin. Oncol. 37, 2368–2377 (2019).
Kimmelman, J., Resnik, D. B., Peppercorn, J. & Ratain, M. J. Burdensome research procedures in trials: why less is more. J. Natl. Cancer Inst. 109, djw315 (2017).
Helft, P. R. & Daugherty, C. K. Are we taking without giving in return? The ethics of research-related biopsies and the benefits of clinical trial participation. J. Clin. Oncol. 24, 4793–4795 (2006).
El-Osta, H. et al. Outcomes of research biopsies in phase I clinical trials: the MD Anderson Cancer Center experience. Oncologist 16, 1292–1298 (2011).
Gomez-Roca, C. A. et al. Sequential research-related biopsies in phase I trials: acceptance, feasibility and safety. Ann. Oncol. 23, 1301–1306 (2012).
Mok, T. S. et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N. Engl. J. Med. 376, 629–640 (2017).
Oxnard, G. R. et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J. Clin. Oncol. 34, 3375–3382 (2016).
Mayrhofer, M. et al. Cell-free DNA profiling of metastatic prostate cancer reveals microsatellite instability, structural rearrangements and clonal hematopoiesis. Genome Med. 10, 85 (2018).
Smith, C. G. et al. Comprehensive characterization of cell-free tumor DNA in plasma and urine of patients with renal tumors. Genome Med. 12, 23 (2020).
Gabriel, E. & Bagaria, S. P. Assessing the impact of circulating tumor DNA (ctDNA) in patients with colorectal cancer: separating fact from fiction. Front. Oncol. 8, 297 (2018).
Miller, A. M. et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature 565, 654–658 (2019).
Li, J. et al. Assessment of ctDNA in CSF may be a more rapid means of assessing surgical outcomes than plasma ctDNA in glioblastoma. Mol. Cell. Probes 46, 101411 (2019).
Micalizzi, D. S., Maheswaran, S. & Haber, D. A. A conduit to metastasis: circulating tumor cell biology. Genes Dev. 31, 1827–1840 (2017).
Rodrigues, P. & Vanharanta, S. Circulating tumor cells: come together, right now, over metastasis. Cancer Discov. 9, 22–24 (2019).
De Rubis, G., Rajeev Krishnan, S. & Bebawy, M. Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharmacol. Sci. 40, 172–186 (2019).
Gast, C. E. et al. Cell fusion potentiates tumor heterogeneity and reveals circulating hybrid cells that correlate with stage and survival. Sci. Adv. 4, eaat7828 (2018).
Miller, M. C., Doyle, G. V. & Terstappen, L. W. M. M. Significance of circulating tumor cells detected by the CellSearch system in patients with metastatic breast colorectal and prostate cancer. J. Oncol. 2010, 617421 (2010).
Bankó, P. et al. Technologies for circulating tumor cell separation from whole blood. J. Hematol. Oncol. 12, 48 (2019).
Scher, H. I. et al. Phenotypic heterogeneity of circulating tumor cells informs clinical decisions between AR signaling inhibitors and taxanes in metastatic prostate cancer. Cancer Res. 77, 5687–5698 (2017).
Stilwell, J. L. et al. RareCyte® CTC analysis step 3: using the CytePicker® module for individual cell retrieval and subsequent whole genome amplification of circulating tumor cells for genomic analysis. Methods Mol. Biol. 1634, 181–192 (2017).
Werbin, J. L. et al. RareCyte® CTC analysis step 2: detection of circulating tumor cells by CyteFinder® automated scanning and semiautomated image analysis. Methods Mol. Biol. 1634, 173–180 (2017).
Ramirez, A. B. et al. RareCyte® CTC analysis step 1: AccuCyte® sample preparation for the comprehensive recovery of nucleated cells from whole blood. Methods Mol. Biol. 1634, 163–172 (2017).
Ohnaga, T., Takei, Y., Nagata, T. & Shimada, Y. Highly efficient capture of cancer cells expressing EGFR by microfluidic methods based on antigen-antibody association. Sci. Rep. 8, 12005 (2018).
Agarwal, A., Balic, M., El-Ashry, D. & Cote, R. J. Circulating tumor cells: strategies for capture, analyses, and propagation. Cancer J. 24, 70–77 (2018).
Cristofanilli, M. et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 351, 781–791 (2004).
De Bono, J. S. et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 14, 6302–6309 (2008).
Cohen, S. J. et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 26, 3213–3221 (2008).
Punnoose, E. A. et al. PTEN loss in circulating tumour cells correlates with PTEN loss in fresh tumour tissue from castration-resistant prostate cancer patients. Br. J. Cancer 113, 1225–1233 (2015).
Ligthart, S. T. et al. Unbiased and automated identification of a circulating tumour cell definition that associates with overall survival. PLoS ONE 6, e27419 (2011).
Werner, S. L. et al. Analytical validation and capabilities of the Epic CTC platform: enrichment-free circulating tumour cell detection and characterization. J. Circ. Biomarkers 4, 3 (2015).
Pailler, E. et al. Acquired resistance mutations to ALK inhibitors identified by single circulating tumor cell sequencing in ALK-rearranged non-small-cell lung cancer. Clin. Cancer Res. 25, 6671–6682 (2019).
Zeune, L. L. et al. How to agree on a CTC: evaluating the consensus in circulating tumor cell scoring. Cytom. Part A 93, 1202–1206 (2018).
Kuhn, P. et al. Lessons learned: transfer of the high-definition circulating tumor cell assay platform to development as a commercialized clinical assay platform. Clin. Pharmacol. Ther. 102, 777–785 (2017).
Kaldjian, E. P. et al. The RareCyte® platform for next-generation analysis of circulating tumor cells. Cytom. Part A 93, 1220–1225 (2018).
Rapanotti, M. C. et al. Minimal residual disease in melanoma: circulating melanoma cells and predictive role of MCAM/MUC18/MelCAM/CD146. Cell Death Discov. 3, 17006 (2017).
Ward, K. et al. Epithelial cell adhesion molecule is expressed in a subset of sarcomas and correlates to the degree of cytological atypia in leiomyosarcomas. Mol. Clin. Oncol. 3, 31–36 (2015).
Sorotsky, H. et al. Quantifying EpCAM heterogeneity of circulating-tumor-cells (CTCs) from small cell lung cancer (SCLC) patients [abstract]. J. Clin. Oncol. 37, e20091 (2019).
Ferreira, M. M., Ramani, V. C. & Jeffrey, S. S. Circulating tumor cell technologies. Mol. Oncol. 10, 374–394 (2016).
Lambros, M. B. et al. Single-cell analyses of prostate cancer liquid biopsies acquired by apheresis. Clin. Cancer Res. 24, 5635–5644 (2018).
Rothwell, D. G. et al. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat. Med. 25, 738–743 (2019).
Sharp, A. et al. Clinical utility of circulating tumour cell androgen receptor splice variant-7 status in metastatic castration-resistant prostate cancer. Eur. Urol. 76, 676–685 (2019).
Sharp, A. et al. Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer. J. Clin. Invest. 129, 192–208 (2019).
Antonarakis, E. S. et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 371, 1028–1038 (2014).
Armstrong, A. J. et al. Prospective multicenter validation of androgen receptor splice variant 7 and hormone therapy resistance in high-risk castration-resistant prostate cancer: the PROPHECY study. J. Clin. Oncol. 37, 1120–1129 (2019).
Plymate, S. R., Sharp, A. & de Bono, J. S. Nuclear circulating tumor cell androgen receptor variant 7 in castration-resistant prostate cancer: the devil is in the detail. JAMA Oncol. 4, 1187–1188 (2018).
Wülfing, P. et al. HER2-positive circulating tumor cells indicate poor clinical outcome in stage I to III breast cancer patients. Clin. Cancer Res. 12, 1715–1720 (2006).
Janning, M. et al. Determination of PD-L1 expression in circulating tumor cells of NSCLC patients and correlation with response to PD-1/PD-L1 inhibitors. Cancers 11, 835 (2019).
Yap, T. A., Lorente, D., Omlin, A., Olmos, D. & De Bono, J. S. Circulating tumor cells: a multifunctional biomarker. Clin. Cancer Res. 20, 2553–2558 (2014).
Smerage, J. B. et al. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: SWOG S0500. J. Clin. Oncol. 32, 3483–3489 (2014).
Marcuello, M. et al. Circulating biomarkers for early detection and clinical management of colorectal cancer. Mol. Aspects Med. 69, 107–122 (2019).
Devriese, L. A., Voest, E. E., Beijnen, J. H. & Schellens, J. H. M. Circulating tumor cells as pharmacodynamic biomarker in early clinical oncological trials. Cancer Treat. Rev. 37, 579–589 (2011).
Siravegna, G., Marsoni, S., Siena, S. & Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 14, 531–548 (2017).
Khetrapal, P. et al. The role of circulating tumour cells and nucleic acids in blood for the detection of bladder cancer: a systematic review. Cancer Treat. Rev. 66, 56–63 (2018).
Garcia-Villa, A. et al. Assessment of γ-H2AX levels in circulating tumor cells from patients receiving chemotherapy. Front. Oncol. 2, 128 (2012).
Pixberg, C. F. et al. Analysis of DNA methylation in single circulating tumor cells. Oncogene 36, 3223–3231 (2017).
Hayes, D. F. et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin. Cancer Res. 12, 4218–4224 (2006).
Budd, G. T. et al. Circulating tumor cells versus imaging – Predicting overall survival in metastatic breast cancer. Clin. Cancer Res. 12, 6403–6409 (2006).
Janni, W. J. et al. Pooled analysis of the prognostic relevance of circulating tumor cells in primary breast cancer. Clin. Cancer Res. 22, 2583–2593 (2016).
Rack, B. et al. Circulating tumor cells predict survival in early average-to-high risk breast cancer patients. J. Natl. Cancer Inst. 106, dju066–11 (2014).
Olmos, D. et al. Baseline circulating tumor cell counts significantly enhance a prognostic score for patients participating in phase I oncology trials. Clin. Cancer Res. 17, 5188–5196 (2011).
Bao, H. et al. Circulating tumor cells: application as a biomarker for molecular characterization and predictor of survival in an all-comer solid tumor phase I clinical study. PLoS ONE 8, e58557 (2013).
Massard, C. et al. RECIST response and variation of circulating tumour cells in phase 1 trials: a prospective multicentric study. Eur. J. Cancer 83, 185–193 (2017).
Heller, G. et al. Circulating tumor cell number as a response measure of prolonged survival for metastatic castration-resistant prostate cancer: a comparison with prostate-specific antigen across five randomized phase III clinical trials. J. Clin. Oncol. 36, 572–580 (2017).
Nagrath, S. et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 450, 1235–1239 (2007).
Chabon, J. J. et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat. Commun. 7, 11815 (2016).
Cremolini, C. et al. Rechallenge for patients with RAS and BRAF wild-type metastatic colorectal cancer with acquired resistance to first-line cetuximab and irinotecan: a phase 2 single-arm clinical trial. JAMA Oncol. 5, 343–350 (2019).
Sorber, L. et al. Circulating cell-free nucleic acids and platelets as a liquid biopsy in the provision of personalized therapy for lung cancer patients. Lung Cancer 107, 100–107 (2017).
Vallée, A. et al. Plasma is a better source of tumor-derived circulating cell-free DNA than serum for the detection of EGFR alterations in lung tumor patients. Lung Cancer 82, 373–374 (2013).
AstraZeneca. IRESSA 250 mg film-coated tablets. Summary of Product Characteristics (AstraZeneca, 2017).
André, F. et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N. Engl. J. Med. 380, 1929–1940 (2019).
US Food and Drug Administration. FDA approves first liquid biopsy next-generation sequencing companion diagnostic test. FDA https://www.fda.gov/news-events/press-announcements/fda-approves-first-liquid-biopsy-next-generation-sequencing-companion-diagnostic-test (2020).
Woodhouse, R. et al. Clinical and analytical validation of FoundationOne liquid CDx, a novel 324-gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS ONE 15, e0237802 (2020).
US Food and Drug Administration. FDA approves liquid biopsy NGS companion diagnostic test for multiple cancers and biomarkers. FDA https://www.fda.gov/drugs/fda-approves-liquid-biopsy-ngs-companion-diagnostic-test-multiple-cancers-and-biomarkers (2021).
Mehra, N. et al. Plasma cell-free DNA concentration and outcomes from taxane therapy in metastatic castration-resistant prostate cancer from two phase III trials (FIRSTANA and PROSELICA). Eur. Urol. 74, 283–291 (2018).
Alix-Panabieres, C. & Pantel, K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 6, 479–491 (2016).
Gupta, R. et al. Guardant360 circulating tumor DNA assay is concordant with FoundationOne next-generation sequencing in detecting actionable driver mutations in anti-EGFR naive metastatic colorectal cancer. Oncologist 25, 235–243 (2020).
Aggarwal, C. et al. Clinical implications of plasma-based genotyping with the delivery of personalized therapy in metastatic non-small cell lung cancer. JAMA Oncol. 5, 173–180 (2019).
Leighl, N. B. et al. Clinical utility of comprehensive cell-free DNA analysis to identify genomic biomarkers in patients with newly diagnosed metastatic non-small cell lung cancer. Clin. Cancer Res. 25, 4691–4700 (2019).
Clark, T. A. et al. Analytical validation of a hybrid capture-based next-generation sequencing clinical assay for genomic profiling of cell-free circulating tumor DNA. J. Mol. Diagn. 20, 686–702 (2018).
Hayes, D. F. et al. Tumor marker utility grading system: a framework to evaluate clinical utility of tumor markers. J. Natl. Cancer Inst. 88, 1456–1466 (1996).
Li, G. et al. Genomic profiling of cell-free circulating tumor DNA in patients with colorectal cancer and its fidelity to the genomics of the tumor biopsy. J. Gastrointest. Oncol. 10, 831–840 (2019).
Mateo, J. et al. DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med. 373, 1697–1708 (2015).
Socinski, M. et al. Final efficacy results from B-F1RST, a prospective phase II trial evaluating blood-based tumour mutational burden (bTMB) as a predictive biomarker for atezolizumab (atezo) in 1L non-small cell lung cancer (NSCLC) [abstract LBA83]. Ann. Oncol. 30 (Suppl. 5), v919–v920 (2019).
Turner, N. C. et al. Circulating tumour DNA analysis to direct therapy in advanced breast cancer (plasmaMATCH): a multicentre, multicohort, phase 2a, platform trial. Lancet Oncol. 21, 1296–1308 (2020).
Gadgeel, S. et al. Phase II/III blood first assay screening trial (BFAST) in patients (pts) with treatment-naïve NSCLC: initial results from the ALK+ cohort [abstract LBA81_PR]. Ann. Oncol. 30 (Suppl. 5), v918 (2019).
Middleton, G. et al. The National Lung Matrix Trial of personalized therapy in lung cancer. Nature 583, 807–812 (2020).
Li, M. M. et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 19, 4–23 (2017).
Remon, J. et al. Liquid biopsy in oncology: a consensus statement of the Spanish Society of Pathology and the Spanish Society of Medical Oncology. Clin. Transl Oncol. 22, 823–834 (2020).
Merker, J. D. et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J. Clin. Oncol. 36, 1631–1641 (2018).
Wu, Y.-M. et al. Inactivation of CDK12 delineates a distinct immunogenic class of advanced prostate cancer. Cell 173, 1770–1782 (2018).
Chen, X. et al. Low-pass whole-genome sequencing of circulating cell-free DNA demonstrates dynamic changes in genomic copy number in a squamous lung cancer clinical cohort. Clin. Cancer Res. 25, 2254–2263 (2019).
Chaudhuri, A. A., Binkley, M. S., Osmundson, E. C., Alizadeh, A. A. & Diehn, M. Predicting radiotherapy responses and treatment outcomes through analysis of circulating tumor DNA. Semin. Radiat. Oncol. 25, 305–312 (2015).
Sumanasuriya, S. et al. Cell-free DNA as a biomarker for taxane treatment in advanced prostate cancer [abstract]. J. Clin. Oncol. 37 (Suppl. 15), 5070 (2019).
Seed, G. et al. Discovery of genomic correlates and tumor purity as an independent clinical factor of poor outcome in advanced prostate cancer lpWGS CNA data [abstract]. Cancer Res. 80 (Suppl. 16), LB-270 (2020).
Dawson, S. J. et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 368, 1199–1209 (2013).
Remon, J. et al. Real-world utility of an amplicon-based next-generation sequencing liquid biopsy for broad molecular profiling in patients with advanced non-small-cell lung cancer. JCO Precis. Oncol. 3, PO.18.00211 (2019).
Sclafani, F. et al. KRAS and BRAF mutations in circulating tumour DNA from locally advanced rectal cancer. Sci. Rep. 8, 1445 (2018).
Perkins, G. et al. Multi-purpose utility of circulating plasma DNA testing in patients with advanced cancers. PLoS ONE 7, e47020 (2012).
Zhou, C. Early clearance of plasma EGFR mutations as a predictor of response to osimertinib and comparator EGFR-TKIs in the FLAURA trial [abstract]. J. Clin. Oncol. 37 (Suppl. 15), 9020 (2020).
Thress, K. S. Complete clearance of plasma EGFR mutations as a predictor of outcome on osimertinib in the AURA trial [abstract]. J. Clin. Oncol. 35 (Suppl. 15), 9018 (2017).
Zhang, Q. et al. Prognostic and predictive impact of circulating tumor DNA in patients with advanced cancers treated with immune checkpoint blockade. Cancer Discov. 10, 1842–1853 (2020).
Goldberg, S. B. et al. Early assessment of lung cancer immunotherapy response via circulating tumor DNA. Clin. Cancer Res. 24, 1872–1880 (2018).
Schiavon, G. et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci. Transl Med. 7, 313ra182 (2015).
Condorelli, R. et al. Polyclonal RB1 mutations and acquired resistance to CDK 4/6 inhibitors in patients with metastatic breast cancer. Ann. Oncol. 29, 640–645 (2018).
Bidard, F.-C. et al. Emergence of ESR1 mutation in cell-free DNA during first line aromatase inhibitor and palbociclib: an exploratory analysis of the PADA-1 trial [abstract]. Ann. Oncol. 30 (Suppl. 5), v105–v106 (2019).
Turner, N. C. et al. Genetic landscape of resistance to CDK4/6 inhibition in circulating tumor DNA (ctDNA) analysis of the PALOMA3 trial of palbociclib and fulvestrant versus placebo and fulvestrant [abstract]. J. Clin. Oncol. 36 (Suppl. 15), 1001 (2018).
O’Leary, B. et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat. Commun. 9, 896 (2018).
Fribbens, C. et al. Tracking evolution of aromatase inhibitor resistance with circulating tumour DNA analysis in metastatic breast cancer. Ann. Oncol. 29, 145–153 (2017).
Lin, K. K. et al. BRCA reversion mutations in circulating tumor DNA predict primary and acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 9, 210–219 (2019).
Khan, K. H. et al. Longitudinal liquid biopsy and mathematical modeling of clonal evolution forecast time to treatment failure in the PROSPECT-C phase II colorectal cancer clinical trial. Cancer Discov. 8, 1270–1285 (2018).
Yang, Z. et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clin. Cancer Res. 24, 3097–3107 (2018).
Shaw, A. T. et al. ALK resistance mutations and efficacy of lorlatinib in advanced anaplastic lymphoma kinase-positive non-small-cell lung cancer. J. Clin. Oncol. 37, 1370–1379 (2019).
Kalluri, R. & LeBleu, V. S. The biology, function, and biomedical applications of exosomes. Science 367, eaau6977 (2020).
Best, M. G. et al. RNA-seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell 28, 666–676 (2015).
Ying, L. et al. Development of a serum miRNA panel for detection of early stage non-small cell lung cancer. Proc. Natl Acad. Sci. USA 117, 25036–25042 (2020).
Kohno, T. et al. Comprehensive analysis of circulating microRNAs as predictive biomarkers for sorafenib therapy outcome in hepatocellular carcinoma. Oncol. Lett. 20, 1727–1733 (2020).
Gioffré, S. et al. Circulating microRNAs as potential predictors of anthracycline-induced troponin elevation in breast cancer patients: diverging effects of doxorubicin and epirubicin. J. Clin. Med. 9, 1418 (2020).
Fan, J. et al. Circulating microRNAs predict the response to anti-PD-1 therapy in non-small cell lung cancer. Genomics 112, 2063–2071 (2020).
Rontogianni, S. et al. Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping. Commun. Biol. 2, 325 (2019).
Rikkert, L. G. et al. Detection of extracellular vesicles in plasma and urine of prostate cancer patients by flow cytometry and surface plasmon resonance imaging. PLoS ONE 15, e0233443 (2020).
de Miguel Pérez, D. et al. Extracellular vesicle-miRNAs as liquid biopsy biomarkers for disease identification and prognosis in metastatic colorectal cancer patients. Sci. Rep. 10, 3974 (2020).
Olmos, D. et al. Prognostic value of blood mRNA expression signatures in castration-resistant prostate cancer: a prospective, two-stage study. Lancet Oncol. 13, 1114–1124 (2012).
Overman, M. J. et al. Use of research biopsies in clinical trials: are risks and benefits adequately discussed? J. Clin. Oncol. 31, 17–22 (2013).
Olson, E. M., Lin, N. U., Krop, I. E. & Winer, E. P. The ethical use of mandatory research biopsies. Nat. Rev. Clin. Oncol. 8, 620–625 (2011).
Hipp, S. et al. A bispecific DLL3/CD3 IgG-Like T-cell engaging antibody induces antitumor responses in small cell lung cancer. Clin. Cancer Res. 26, 5258–5268 (2020).
Puca, L. et al. Delta-like protein 3 expression and therapeutic targeting in neuroendocrine prostate cancer. Sci. Transl Med. 11, eaav0891 (2019).
Acknowledgements
P.W. acknowledges funding from Cancer Research UK (C309/A11566 and C35696/A23187), Wellcome (212969/Z/18/Z) and the Mark Foundation and the Chordoma Foundation. J.d.B. acknowledges funding support from Cancer Research UK through The ICR Cancer Centre Grant, from an Experimental Cancer Medicines Centre grant funded by Cancer Research UK and the Department of Health to The Institute of Cancer Research and Royal Marsden, from Movember to the London Prostate Cancer Centre of Excellence programme, and from Prostate Cancer UK and the Prostate Cancer Foundation.
Author information
Authors and Affiliations
Contributions
All authors made substantial contributions to each stage of the preparation of this manuscript.
Corresponding author
Ethics declarations
Competing interests
M.S.M. has become an employee of AstraZeneca since this manuscript was initiated, and is a current shareholder of this company; however, this affiliation does not lead to a direct conflict of interest with the presented work. P.W. is a consultant and/or scientific advisory board member for Astex Pharmaceuticals, Black Diamond Therapeutics, CV6, NextechInvest and Storm Therapeutics, and holds stock in Black Diamond Therapeutics, Chroma Therapeutics, NextechInvest and Storm Therapeutics; he is also a former employee of AstraZeneca and a Non-Executive Director of Storm Therapeutics and the Royal Marsden NHS Trust, and is an Executive Director of the non-profit Chemical Probes Portal. P.W. has received research funding from CRT Pioneer Fund/6th Element Capital and Merck. J.d.B. has served on advisory boards for Astellas, AstraZeneca, Bayer, Boehringer Ingelheim, Daiichi, Eisai, Genentech/Roche, Genmab, GlaxoSmithKline, Janssen, Merck Serono, Merck Sharp & Dohme, Menarini Silicon Biosystems, Orion, Pfizer, Qiagen, Sanofi-Aventis, Sierra Oncology, Taiho and Vertex Pharmaceuticals. He is an employee of The Institute of Cancer Research, which has received funding or other support for his research work from Astellas, AstraZeneca, Bayer, Cellcentric, Daiichi, Genentech, Genmab, GlaxoSmithKline, Janssen, Merck Serono, Merck Sharp & Dohme, Menarini Silicon Biosystems, Orion, Pfizer, Sanofi-Aventis, Sierra Oncology, Taiho and Vertex. The Institute of Cancer Research also has a commercial interest in abiraterone, PARP inhibition in DNA repair-defective cancers and PI3K/AKT pathway inhibitors (which provide no personal income to J.d.B.). J.d.B. is named as an inventor, with no financial interest, on US patent 8,822,438. A.P. and R.S. declare no competing interests.
Additional information
Peer review information
Nature Reviews Clinical Oncology thanks J.-Y. Pierga, C. Rolfo, A. Sartore-Bianchi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Pal, A., Shinde, R., Miralles, M.S. et al. Applications of liquid biopsy in the Pharmacological Audit Trail for anticancer drug development. Nat Rev Clin Oncol 18, 454–467 (2021). https://doi.org/10.1038/s41571-021-00489-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41571-021-00489-x
This article is cited by
-
The development of a novel zeolite-based assay for efficient and deep plasma proteomic profiling
Journal of Nanobiotechnology (2024)