Abstract
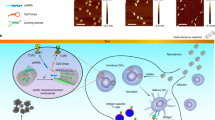
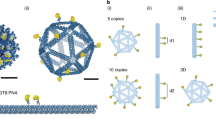
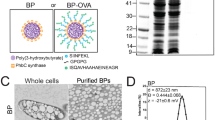
Multivalent presentation of ligands often enhances receptor activation and downstream signalling. DNA origami offers a precise nanoscale spacing of ligands, a potentially useful feature for therapeutic nanoparticles. Here we use a square-block DNA origami platform to explore the importance of the spacing of CpG oligonucleotides. CpG engages Toll-like receptors and therefore acts to activate dendritic cells. Through in vitro cell culture studies and in vivo tumour treatment models, we demonstrate that square blocks induce Th1 immune polarization when CpG is spaced at 3.5 nm. We observe that this DNA origami vaccine enhances DC activation, antigen cross-presentation, CD8 T-cell activation, Th1-polarized CD4 activation and natural-killer-cell activation. The vaccine also effectively synergizes with anti-PD-L1 for improved cancer immunotherapy in melanoma and lymphoma models and induces long-term T-cell memory. Our results suggest that DNA origami may serve as a platform for controlling adjuvant spacing and co-delivering antigens in vaccines.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Data supporting the findings of this study are available in the Article and its Supplementary Information. The RNAseq data for the stimulated BMDCs generated and analysed in this Article have been deposited in NCBI’s Gene Expression Omnibus under accession no. GSE251850. Source data are provided with this paper.
References
Sahin, U. & Tureci, O. Personalized vaccines for cancer immunotherapy. Science 359, 1355–1360 (2018).
Bauer, S. et al. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc. Natl Acad. Sci. USA 98, 9237–9242 (2001).
Bode, C., Zhao, G., Steinhagen, F., Kinjo, T. & Klinman, D. M. CpG DNA as a vaccine adjuvant. Expert Rev. Vaccines 10, 499–511 (2011).
Klinman, D. M., Sato, T. & Shimosato, T. Use of nanoparticles to deliver immunomodulatory oligonucleotides. WIREs Nanomed. Nanobiotechnol. 8, 631–637 (2016).
Schuller, V. J. et al. Cellular immunostimulation by CpG-sequence-coated DNA origami structures. ACS Nano 5, 9696–9702 (2011).
Casaletto, J. B. & McClatchey, A. I. Spatial regulation of receptor tyrosine kinases in development and cancer. Nat. Rev. Cancer 12, 387–400 (2012).
Shaw, A. et al. Spatial control of membrane receptor function using ligand nanocalipers. Nat. Methods 11, 841–846 (2014).
Kwon, P. S. et al. Designer DNA architecture offers precise and multivalent spatial pattern-recognition for viral sensing and inhibition. Nat. Chem. 12, 26–35 (2020).
Pulendran, B. & Ahmed, R. Translating innate immunity into immunological memory: implications for vaccine development. Cell 124, 849–863 (2006).
Ohto, U. et al. Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9. Nature 520, 702–705 (2015).
Leleux, J. A., Pradhan, P. & Roy, K. Biophysical attributes of CpG presentation control TLR9 signaling to differentially polarize systemic immune responses. Cell Rep. 18, 700–710 (2017).
Schmidt, N. W. et al. Liquid-crystalline ordering of antimicrobial peptide-DNA complexes controls TLR9 activation. Nat. Mater. 14, 696–700 (2015).
Lee, E. Y. et al. A review of immune amplification via ligand clustering by self-assembled liquid-crystalline DNA complexes. Adv. Colloid Interface Sci. 232, 17–24 (2016).
Comberlato, A., Koga, M. M., Nussing, S., Parish, I. A. & Bastings, M. M. C. Spatially controlled activation of Toll-like receptor 9 with DNA-based nanomaterials. Nano Lett. 22, 2506–2513 (2022).
Du, R. R. et al. Innate immune stimulation using 3D wireframe DNA origami. ACS Nano 16, 20340–20352 (2022).
Johansson, M., Denardo, D. G. & Coussens, L. M. Polarized immune responses differentially regulate cancer development. Immunol. Rev. 222, 145–154 (2008).
Yew, N. S. et al. CpG-depleted plasmid DNA vectors with enhanced safety and long-term gene expression in vivo. Mol. Ther. 5, 731–738 (2002).
Kumar, V. et al. DNA nanotechnology for cancer therapy. Theranostics 6, 710–725 (2016).
Udomprasert, A. & Kangsamaksin, T. DNA origami applications in cancer therapy. Cancer Sci. 108, 1535–1543 (2017).
Li, S. et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 36, 258–264 (2018).
Liu, S. et al. A DNA nanodevice-based vaccine for cancer immunotherapy. Nat. Mater. 20, 421–430 (2021).
Kern, N., Dong, R., Douglas, S. M., Vale, R. D. & Morrissey, M. A. Tight nanoscale clustering of Fcγ receptors using DNA origami promotes phagocytosis. eLife 10, e68311 (2021).
Berger, R. M. L. et al. Nanoscale FasL organization on DNA origami to decipher apoptosis signal activation in cells. Small 17, e2101678 (2021).
Rothemund, P. W. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Douglas, S. M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Liedl, T., Hogberg, B., Tytell, J., Ingber, D. E. & Shih, W. M. Self-assembly of three-dimensional prestressed tensegrity structures from DNA. Nat. Nanotechnol. 5, 520–524 (2010).
Shih, W. M. Exploiting weak interactions in DNA self-assembly. Science 347, 1417–1418 (2015).
Dietz, H., Douglas, S. M. & Shih, W. M. Folding DNA into twisted and curved nanoscale shapes. Science 325, 725–730 (2009).
Shen, H. et al. Enhanced and prolonged cross-presentation following endosomal escape of exogenous antigens encapsulated in biodegradable nanoparticles. Immunology 117, 78–88 (2006).
Min, Y. et al. Antigen-capturing nanoparticles improve the abscopal effect and cancer immunotherapy. Nat. Nanotechnol. 12, 877–882 (2017).
Chesson, C. B. & Zloza, A. Nanoparticles: augmenting tumor antigen presentation for vaccine and immunotherapy treatments of cancer. Nanomedicine 12, 2693–2706 (2017).
Ponnuswamy, N. et al. Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nat. Commun. 8, 15654 (2017).
Anastassacos, F. M., Zhao, Z., Zeng, Y. & Shih, W. M. Glutaraldehyde cross-linking of oligolysines coating DNA origami greatly reduces susceptibility to nuclease degradation. J. Am. Chem. Soc. 142, 3311–3315 (2020).
Lucas, C. R. et al. DNA origami nanostructures elicit dose-dependent immunogenicity and are nontoxic up to high doses in vivo. Small 18, e2108063 (2022).
Wamhoff, E. C. et al. Evaluation of nonmodified wireframe DNA origami for acute toxicity and biodistribution in mice. ACS Appl. Bio. Mater. 6, 1960–1969 (2023).
Douglas, S. M. et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 37, 5001–5006 (2009).
Njongmeta, L. M. et al. CD205 antigen targeting combined with dendritic cell recruitment factors and antigen-linked CD40L activation primes and expands significant antigen-specific antibody and CD4(+) T cell responses following DNA vaccination of outbred animals. Vaccine 30, 1624–1635 (2012).
Lahoud, M. H. et al. DEC-205 is a cell surface receptor for CpG oligonucleotides. Proc. Natl Acad. Sci. USA 109, 16270–16275 (2012).
You, C. X. et al. AAV2/IL-12 gene delivery into dendritic cells (DC) enhances CTL stimulation above other IL-12 applications: evidence for IL-12 intracrine activity in DC. Oncoimmunology 1, 847–855 (2012).
Heo, M. B., Kim, S. Y., Yun, W. S. & Lim, Y. T. Sequential delivery of an anticancer drug and combined immunomodulatory nanoparticles for efficient chemoimmunotherapy. Int J. Nanomed. 10, 5981–5992 (2015).
Scheuerpflug, A. et al. The role of dendritic cells for therapy of B-cell lymphoma with immune checkpoint inhibitors. Cancer Immunol. Immunother. 70, 1343–1350 (2020).
Keestra, A. M., de Zoete, M. R., Bouwman, L. I. & van Putten, J. P. Chicken TLR21 is an innate CpG DNA receptor distinct from mammalian TLR9. J. Immunol. 185, 460–467 (2010).
Oldenburg, M. et al. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science 337, 1111–1115 (2012).
Spies, B. et al. Vaccination with plasmid DNA activates dendritic cells via Toll-like receptor 9 (TLR9) but functions in TLR9-deficient mice. J. Immunol. 171, 5908–5912 (2003).
Yu, D. et al. ‘Immunomers’–novel 3′-3′-linked CpG oligodeoxyribonucleotides as potent immunomodulatory agents. Nucleic Acids Res. 30, 4460–4469 (2002).
Minari, J., Mochizuki, S. & Sakurai, K. Enhanced cytokine secretion owing to multiple CpG side chains of DNA duplex. Oligonucleotides 18, 337–344 (2008).
Smith, L. K. et al. Interleukin-10 directly inhibits CD8(+) T cell function by enhancing N-glycan branching to decrease antigen sensitivity. Immunity 48, 299–312 e295 (2018).
Li, A. W. et al. A facile approach to enhance antigen response for personalized cancer vaccination. Nat. Mater. 17, 528–534 (2018).
Kreiter, S. et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 520, 692–696 (2015).
Toubi, E. & Shoenfeld, Y. Protective autoimmunity in cancer (review). Oncol. Rep. 17, 245–251 (2007).
Ke, Y., Voigt, N. V., Gothelf, K. V. & Shih, W. M. Multilayer DNA origami packed on hexagonal and hybrid lattices. J. Am. Chem. Soc. 134, 1770–1774 (2012).
Douglas, S. M., Chou, J. J. & Shih, W. M. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc. Natl Acad. Sci. USA 104, 6644–6648 (2007).
Hahn, J., Wickham, S. F., Shih, W. M. & Perrault, S. D. Addressing the instability of DNA nanostructures in tissue culture. ACS Nano 8, 8765–8775 (2014).
Acknowledgements
We thank Q. Yan, Z. Zhao, T. Zhang, P. Lill, P. Prabhala, L. Chou, J. Han, D. Minev, B. Everhart, J. Deng, D. Zhang, K. Adu-Berchie, H. Dembele, A. Rajwar and K. Simpson for aiding in labour support, experimental design, exploring experiments and manuscript proofreading. We also thank M. Perez, M. Carr and E. Zigon for their assistance in lab management and facility usage. We thank M. Bastings and A. Li for exploration—before the current project initiated—of CpG-functionalized DNA origami barrels for immune stimulation. This work was funded by the Barr Award granted by the Claudia Adams Barr Program (Y.C.Z.) in the Dana-Farber Cancer Institute and by Wyss validation funding (Y.C.Z.) at the Wyss Institute for Biologically Inspired Engineering at Harvard. This study was supported by the Korean Fund for Regenerative Medicine (J.H.R.; 21A0504L1) funded by the Korean government (the Ministry of Science and ICT and the Korean Ministry of Health & Welfare) and the Intramural Research Program of KIST (J.H.R.; no. 2E30840). This project was also supported by an NIH U54 grant (W.M.S.; CA244726-01).
Author information
Authors and Affiliations
Contributions
Y.C.Z. developed and planned the experiments, carried out the vaccine fabrication and validation, and wrote the manuscript. J.H.R. and W.M.S. provided the experimental and theoretical guidance and edited the manuscript. O.J.Y. assisted Y.C.Z. in performing the experiments, analysing the data and manuscript editing. C.M.W., F.M.A., J.I.M. and G.I. assisted with the DNA origami design, modelling and fabrication. H.B. performed the RNAseq analysis. M.O.D., M. Sobral, M. Sanchez, A.R.G. and A.V. helped with the animal study design, modelling and sampling. T.C.F. assisted with the confocal experiment. Y.C. assisted with the 3D modelling and manuscript editing. G.G.F. and D.N. guided the statistical analysis. C.J.W. and D.B.K. guided the experiment design and offered manuscript editing. D.J.M. and I.C.K. provided project support and manuscript editing.
Corresponding authors
Ethics declarations
Competing interests
W.M.S., J.H.R. and Y.C.Z. are inventors on US patent application PCT/US2020/036281 held by Dana-Farber Cancer Institute, Korea Institute of Science and Technology and Wyss Institute (filed on 5 June 2020) partially based on this work. All other authors declare no competing interests.
Peer review
Peer review information
Nature Nanotechnology thanks Mark Bathe, Diana Morzy and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–34, Tables 1–6, Discussion and Methods.
Supplementary Information
Supplementary Video 1 This video demonstrates the effective uptake of DNA origami nanoparticles coated with K10-PEG5k by DCs within a time frame of 2 h and reveals the presence of DNA origami nanoparticles (pink) conjugated with OVA antigen (green) within the DCs, along with late endosomes (cyan).
Supplementary Data 1
Statistical source data.
Supplementary Data 2
Statistical source data.
Supplementary Data 3
Statistical source data.
Supplementary Data 4
Statistical source data.
Supplementary Data 5
Statistical source data.
Supplementary Data 6
Statistical source data.
Supplementary Data 7
Statistical source data.
Supplementary Data 8
Statistical source data.
Supplementary Data 9
Statistical source data.
Supplementary Data 10
Statistical source data.
Supplementary Data 11
Statistical source data.
Supplementary Data 12
Statistical source data.
Supplementary Data 13
Statistical source data.
Supplementary Data 14
Statistical source data.
Supplementary Data 15
Statistical source data.
Supplementary Data 16
Statistical source data.
Supplementary Data 17
Statistical source data.
Supplementary Data 18
Statistical source data.
Supplementary Data 19
Statistical source data.
Supplementary Data 20
Statistical source data.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zeng, Y.C., Young, O.J., Wintersinger, C.M. et al. Fine tuning of CpG spatial distribution with DNA origami for improved cancer vaccination. Nat. Nanotechnol. (2024). https://doi.org/10.1038/s41565-024-01615-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41565-024-01615-3