Abstract
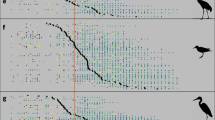
Climate change, land-use change, pollution and exploitation are among the main drivers of species’ population trends; however, their relative importance is much debated. We used a unique collection of over 1,000 local population time series in 22 communities across terrestrial, freshwater and marine realms within central Europe to compare the impacts of long-term temperature change and other environmental drivers from 1980 onwards. To disentangle different drivers, we related species’ population trends to species- and driver-specific attributes, such as temperature and habitat preference or pollution tolerance. We found a consistent impact of temperature change on the local abundances of terrestrial species. Populations of warm-dwelling species increased more than those of cold-dwelling species. In contrast, impacts of temperature change on aquatic species’ abundances were variable. Effects of temperature preference were more consistent in terrestrial communities than effects of habitat preference, suggesting that the impacts of temperature change have become widespread for recent changes in abundance within many terrestrial communities of central Europe.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Butchart, S. H. M. et al. Global biodiversity: indicators of recent declines. Science 328, 1164–1168 (2010).
Living Planet Report 2016: Risk and Resilience in a New Era (WWF International, 2016).
Winfree, R., Fox, J. W., Williams, N. M., Reilly, J. R. & Cariveau, D. P. Abundance of common species, not species richness, drives delivery of a real-world ecosystem service. Ecol. Lett. 18, 626–635 (2015).
Mair, L. et al. Abundance changes and habitat availability drive species’ responses to climate change. Nat. Clim. Change 4, 127–131 (2014).
Johnston, A. et al. Observed and predicted effects of climate change on species abundance in protected areas. Nat. Clim. Change 3, 1055–1061 (2013).
Urban, M. C. Accelerating extinction risk from climate change. Science 348, 571–573 (2015).
Chen, I. C., Hill, J. K., Ohlemuller, R., Roy, D. B. & Thomas, C. D. Rapid range shifts of species associated with high levels of climate warming. Science 333, 1024–1026 (2011).
Gillings, S., Balmer, D. E. & Fuller, R. J. Directionality of recent bird distribution shifts and climate change in Great Britain. Glob. Change Biol. 21, 2155–2168 (2015).
Virkkala, R. & Lehikoinen, A. Patterns of climate-induced density shifts of species: poleward shifts faster in northern boreal birds than in southern birds. Glob. Change Biol. 20, 2995–3003 (2014).
Devictor, V. et al. Differences in the climatic debts of birds and butterflies at a continental scale. Nat. Clim. Change 2, 121–124 (2012).
Sala, O. E. et al. Biodiversity—global biodiversity scenarios for the year 2100. Science 287, 1770–1774 (2000).
Conti, M. E. & Cecchetti, G. Biological monitoring: lichens as bioindicators of air pollution assessment—a review. Environ. Pollut. 114, 471–492 (2001).
Donald, P. F., Green, R. E. & Heath, M. F. Agricultural intensification and the collapse of Europe’s farmland bird populations. Proc. R. Soc. Lond. B 268, 25–29 (2001).
Burns, F. et al. Agricultural management and climatic change are the major drivers of biodiversity change in the UK. PLoS ONE 11, e0151595 (2016).
Adrian, R., Gerten, D., Huber, V., Wagner, C. & Schmidt, S. R. Windows of change: temporal scale of analysis is decisive to detect ecosystem responses to climate change. Mar. Biol. 159, 2533–2542 (2012).
Guisan, A. & Thuiller, W. Predicting species distribution: offering more than simple habitat models. Ecol. Lett. 8, 993–1009 (2005); erratum 10, 435–435 (2007).
Parmesan, C. et al. Beyond climate change attribution in conservation and ecological research. Ecol. Lett. 16, 58–71 (2013).
Williams, S. E., Shoo, L. P., Isaac, J. L., Hoffmann, A. A. & Langham, G. Towards an integrated framework for assessing the vulnerability of species to climate change. PLoS Biol. 6, 2621–2626 (2008).
van Herk, C. M., Aptroot, A. & van Dobben, H. F. Long-term monitoring in the Netherlands suggests that lichens respond to global warming. Lichenologist 34, 141–154 (2002).
Cahill, A. E. et al. How does climate change cause extinction? Proc. R. Soc. B 280, 20121890 (2013).
Seppelt, R., Manceur, A. M., Liu, J. G., Fenichel, E. P. & Klotz, S. Synchronized peak-rate years of global resources use. Ecol. Soc. 19, 50 (2014).
Eglington, S. M. & Pearce-Higgins, J. W. Disentangling the relative importance of changes in climate and land-use intensity in driving recent bird population trends. PLoS ONE 7, e30407 (2012).
van Swaay, C., Warren, M. & Lois, G. Biotope use and trends of European butterflies. J. Insect Conserv. 10, 189–209 (2006).
Venter, O. et al. Sixteen years of change in the global terrestrial human footprint and implications for biodiversity conservation. Nat. Commun. 7, 12558 (2016).
Simpson, S. D. et al. Continental shelf-wide response of a fish assemblage to rapid warming of the sea. Curr. Biol. 21, 1565–1570 (2011).
Daufresne, M. & Boet, P. Climate change impacts on structure and diversity of fish communities in rivers. Glob. Change Biol. 13, 2467–2478 (2007).
Floury, M., Usseglio-Polatera, P., Ferreol, M., Delattre, C. & Souchon, Y. Global climate change in large European rivers: long-term effects on macroinvertebrate communities and potential local confounding factors. Glob. Change Biol. 19, 1085–1099 (2013).
Jeppesen, E. et al. Impacts of climate warming on the long-term dynamics of key fish species in 24 European lakes. Hydrobiologia 694, 1–39 (2012).
Wagner, C. & Adrian, R. Consequences of changes in thermal regime for plankton diversity and trait composition in a polymictic lake: a matter of temporal scale. Freshwat. Biol. 56, 1949–1961 (2011).
Devictor, V., Julliard, R., Couvet, D. & Jiguet, F. Birds are tracking climate warming, but not fast enough. Proc. R. Soc. B 275, 2743–2748 (2008).
Clavero, M., Villero, D. & Brotons, L. Climate change or land use dynamics: do we know what climate change indicators indicate? PLoS ONE 6, e18581 (2011).
Guisan, A. & Thuiller, W. Predicting species distribution: offering more than simple habitat models. Ecol. Lett. 8, 993–1009 (2005).
Storlie, C. et al. Stepping inside the niche: microclimate data are critical for accurate assessment of species’ vulnerability to climate change. Biol. Lett. 10, 20140576 (2014).
Homburg, K., Homburg, N., Schaefer, F., Schuldt, A. & Assmann, T. Carabids. org—A dynamic online database of ground beetle species traits (Coleoptera, Carabidae). Insect Conserv. Div. 7, 195–205 (2014).
Burkhardt, U. et al. The Edaphobase project of GBIF-Germany—a new online soil-zoological data warehouse. Appl. Soil Ecol. 83, 3–12 (2014).
Poloczanska, E. S. et al. Global imprint of climate change on marine life. Nat. Clim. Change 3, 919–925 (2013).
Rue, H., Martino, S. & Chopin, N. Approximate Bayesian inference for latent Gaussian models by using integrated nested Laplace approximations. J. R. Stat. Soc. B 71, 319–392 (2009).
Haylock, M. R. et al. A European daily high-resolution gridded data set of surface temperature and precipitation for 1950–2006. J. Geophys. Res. 113, D20119 (2008).
Caissie, D. The thermal regime of rivers: a review. Freshwat. Biol. 51, 1389–1406 (2006).
Kaschner, K. et al. AquaMaps Environmental Dataset: Half-Degree Cells Authority File (HCAF) v. 07/2010 (AquaMaps, accessed 25 July 2013); www.aquamaps.org/data
Grewe, Y., Hof, C., Dehling, D. M., Brandl, R. & Brandle, M. Recent range shifts of European dragonflies provide support for an inverse relationship between habitat predictability and dispersal. Glob. Ecol. Biogeogr. 22, 403–409 (2013).
Devictor, V., Julliard, R. & Jiguet, F. Distribution of specialist and generalist species along spatial gradients of habitat disturbance and fragmentation. Oikos 117, 507–514 (2008).
Penone, C. et al. Imputation of missing data in life-history trait datasets: which approach performs the best? Methods Ecol. Evol. 5, 961–970 (2014).
Koller, M. & Stahel, W. A. Sharpening Wald-type inference in robust regression for small samples. Comput. Stat. Data Anal. 55, 2504–2515 (2011).
Domisch, S. et al. Modelling distribution in European stream macroinvertebrates under future climates. Glob. Change Biol. 19, 752–762 (2013).
Jetz, W., Thomas, G. H., Joy, J. B., Hartmann, K. & Mooers, A. O. The global diversity of birds in space and time. Nature 491, 444–448 (2012).
Drummond, A. J., Suchard, M. A., Xie, D. & Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (2012).
Fritz, S. A., Bininda-Emonds, O. R. P. & Purvis, A. Geographical variation in predictors of mammalian extinction risk: big is bad, but only in the tropics. Ecol. Lett. 12, 538–549 (2009).
Durka, W. & Michalski, S. G. Daphne: a dated phylogeny of a large European flora for phylogenetically informed ecological analyses. Ecology 93, 2297–2297 (2012).
Dunger, W. & Burkhardt, U. Synopses on Palearctic Collembola (Museum Natural History, Görlitz, 2012).
Guiry, M. D. & Guiry, G. M. AlgaeBase (National University of Ireland, 2017); http://www.algaebase.org
Münkemüller, T. et al. How to measure and test phylogenetic signal. Methods Ecol. Evol. 3, 743–756 (2012).
Jombart, T., Balloux, F. & Dray, S. Adephylo: new tools for investigating the phylogenetic signal in biological traits. Bioinformatics 26, 1907–1909 (2010).
Paradis, E., Claude, J. & Strimmer, K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004).
Nakagawa, S. & Cuthill, I. C. Effect size, confidence interval and statistical significance: a practical guide for biologists. Biol. Rev. 82, 591–605 (2007).
Koricheva, J., Gurevitch, J. & Mengersen, K. Handbook of Meta-analysis in Ecology and Evolution (Princeton Univ. Press, 2013).
R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2013).
Acknowledgements
We thank Bayerisches Landesamt für Umwelt, Sächsisches Landesamt für Umwelt, Landwirtschaft und Geologie, Landesanstalt für Umwelt, Messungen und Naturschutz Baden-Württemberg, Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen, Hessisches Landesamt für Umwelt und Geologie and the Trilateral Monitoring and Assessment Program (TMAP) for sharing and providing permission to use their data for our project. Additionally, we appreciate the open access marine data provided by the International Council for the Exploration of the Sea. We thank the following scientists for taxonomic or technical advice: C. Brendel, T. Caprano, R. Claus, K. Desender, A. Flakus, P. R. Flakus, S. Fritz, E.-M. Gerstner, J.-P. Maelfait, E.-L. Neuschulz, S. Pauls, C. Printzen, I. Schmitt and H. Turin, and I. Bartomeus for comments on a previous version of the manuscript. R.A. was supported by the EU-project LIMNOTIP funded under the seventh European Commission Framework Programme (FP7) ERA-Net Scheme (Biodiversa, 01LC1207A) and the long-term ecological research program at the Leibniz-Institute of Freshwater Ecology and Inland Fisheries (IGB). R.W.B. was supported by the Scottish Government Rural and Environment Science and Analytical Services Division (RESAS) through Theme 3 of their Strategic Research Programme. S.D. acknowledges support of the German Research Foundation DFG (grant DO 1880/1-1). S.S. acknowledges the support from the FP7 project EU BON (grant no. 308454). S.K., I.Kü. and O.S. acknowledge funding thorough the Helmholtz Association’s Programme Oriented Funding, Topic ‘Land use, biodiversity, and ecosystem services: Sustaining human livelihoods’. O.S. also acknowledges the support from FP7 via the Integrated Project STEP (grant no. 244090). D.E.B. was funded by a Landes–Offensive zur Entwicklung Wissenschaftlich–ökonomischer Exzellenz (LOEWE) excellence initiative of the Hessian Ministry for Science and the Arts and the German Research Foundation (DFG: Grant no. BO 1221/23-1).
Author information
Authors and Affiliations
Contributions
D.E.B. performed the analysis and wrote the outline of the paper with K.B.G. The study and analysis was perceived and designed by D.E.B., C.H., P.H., I.Kr., O.S. and K.B.G. All remaining authors contributed data towards the analysis. All authors helped draft the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1–12; Supplementary Tables 1–5 (PDF 2190 kb)
Rights and permissions
About this article
Cite this article
Bowler, D., Hof, C., Haase, P. et al. Cross-realm assessment of climate change impacts on species’ abundance trends. Nat Ecol Evol 1, 0067 (2017). https://doi.org/10.1038/s41559-016-0067
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41559-016-0067
This article is cited by
-
Trait-mediated processes and per capita contributions to ecosystem functioning depend on conspecific density and climate conditions
Communications Earth & Environment (2024)
-
Warming underpins community turnover in temperate freshwater and terrestrial communities
Nature Communications (2024)
-
Analysis of the state of conservation of Trachurus trachurus in the Western Mediterranean Sea based on the interannual (2009–2020) changes in their life history traits
Marine Biology (2024)
-
Mechanisms, detection and impacts of species redistributions under climate change
Nature Reviews Earth & Environment (2024)
-
Global patterns of climate change impacts on desert bird communities
Nature Communications (2023)