Abstract
Purpose
Patients with reciprocal balanced translocations (RBT) have a risk for recurrent pregnancy losses (RPL), affected child, and infertility. Currently, genetic counseling is based on karyotypes found among the products of conception (POC), although factors influencing the success of assisted reproductive technologies (ART) in RBT couples are not established.
Methods
Cytogenetic results from 261 POC and offspring of the parents (113 women and 90 men) with RBT were evaluated. Chromosome segregation modes and number of euploid embryos were assessed in couples undergoing in vitro fertilization.
Results
Patients with translocations involving an acrocentric chromosome have a higher risk of unbalanced gametes caused by a 3:1 segregation. Female RBT patients have a statistically higher risk of aneuploidy due to an interchromosomal effect. The rate of euploid embryos is low due to meiosis I malsegregation of RBT, meiosis II nondisjunction, additional whole chromosome or segmental aneusomies. RBT patients with RPL have a higher rate of miscarriage of euploid fetuses with RBT.
Conclusion
Chromosome-specific factors, female gender, age, and history of RPL are the risk elements influencing pregnancy and in vitro fertilization success in RBT patients. Chromosomal microarray analysis of POC is necessary to provide an accurate and timely diagnosis for patients with adverse reproductive outcomes.
Similar content being viewed by others
INTRODUCTION
Reciprocal translocations are chromosome aberrations characterized by an exchange of DNA segments between nonhomologous chromosomes with no gain or loss of DNA at the breakpoints and are thus balanced rearrangements. Individuals with reciprocal balanced translocations (RBT) are usually phenotypically normal unless the translocation breakpoints disrupt a dominant gene(s) or the transposition of chromosomal segments has an effect on the expression of nearby genes. Near 27% of patients with a de novo RBT have birth defects and/or neurodevelopmental disorders,1 while all individuals with translocations are at risk of reproductive failures.
Balanced translocations are diagnosed following cytogenetic testing due to reproductive issues such as infertility, repeated pregnancy losses (RPL), stillbirths, or the birth of a child with congenital anomalies and an unbalanced karyotype. The estimated frequency of RBT is ~1/500 (~0.15–0.2%) in the general population.2,3 In infertile males, the frequency of translocations is ~1.3%,4,5 and in 6.6% of couples with RPL, one of the partners has a RBT.6,7,8
The RBT incidence is likely underestimated, as classical G-banding karyotype analysis will yield normal results in individuals with submicroscopic chromosome rearrangements. Multiple studies have demonstrated the utility of chromosomal microarray analysis (CMA) in detection of genomic imbalances that might be present in fetuses or products of conception (POC).9,10,11,12,13 The application of CMA to POC for couples with RPL is still limited, reducing the likelihood of accurate diagnosis of parental RBT.
In gametogenesis, chromosomal segregation of translocated chromosomes can produce balanced and unbalanced gametes. During meiosis I, the translocated chromosomes and their normal homologues form a quadrivalent structure that undergoes segregation via five possible modes: alternate, adjacent-1, adjacent-2, 3:1, or 4:0 (Fig. 1a). Alternate segregation results in the production of only balanced gametes. Adjacent-1, adjacent-2, 3:1, and 4:0 segregations will produce unbalanced gametes. The segregation mode in RBT patients is difficult to predict as it is influenced by many factors, including the structure and size of a chromosome involved in a translocation, the size of the translocated segments, unique position of the translocation breakpoints in each patient, gene content, patient’s gender, and other aspects.
(a) Possible chromosomal complements in gametes of patients with balanced translocations. At meiosis I, translocated chromosomes and their normal homologues synapse to form a quadrivalent. Balanced gametes containing either the normal nonhomologous chromosomes or both translocated chromosomes produced by alternate segregation are specified by green borders. Unbalanced gametes are specified by red borders. Modes of chromosome segregation from a quadrivalent are shown: 2:2 (two nonhomologous or two homologous chromosomes segregate together in adjacent-1 or adjacent-2 disjunction, respectively), 3:1 (three chromosomes segregate to one cell and one to the other), and 4:0 (all chromosomes segregate together). †Nondisjunction of the sister chromatids during meiosis II may result in occurrence of gametes with additional chromosome abnormalities. (b) Pie chart showing reproductive outcomes. (c) The rate of aneuploid conceptions in male and female patients. Asterisks indicate statistically significant sex bias. No difference in male and female patients was observed for 3:1 segregation mode.
The effect of a translocation on fertility and pregnancy outcomes is unknown for the majority of patients. More reliable data are available on recurrent translocations, such as t(11;22)(q23;q11),14 and Robertsonian translocations, a nonreciprocal rearrangement resulting in a fusion of the long arms of two acrocentric chromosomes and formation of a single chromosome, such as der(14;21)(q10;q10).
Segregation events producing unbalanced chromosomal complements will remain largely unidentified if they result in infertility or early embryonic/fetal demise. Studies in couples opting for in vitro fertilization (IVF) and preimplantation genetic testing for aneuploidies (PGT-A) or structural rearrangements (PGT-SR) identify that RBT patients produce ~10–20% of euploid embryos.15,16 Besides the risk of unbalanced gametes and an age-related aneuploidy, RBT individuals may have additional risk factors diminishing the number of euploid embryos available for implantation. In this study, we analyzed the reproductive outcomes in 203 couples with RBT, evaluated sex-biased differences of RBT patients on meiotic segregations and incidence of a concurrent aneuploidy, and defined chromosome-specific factors that may influence successful IVF.
MATERIALS AND METHODS
Study population
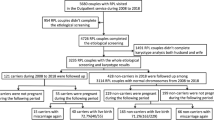
The study was approved by the University of Pittsburgh institutional review board (IRB) (STUDY20060192; STUDY18120172; STUDY20050099). We assessed cytogenetic abnormalities and reproductive outcomes in 203 couples (113 women and 90 men) carrying RBT. Robertsonian translocations were excluded. Retrospective review of medical records was performed to obtain couples’ reproductive history, cytogenetic results, and outcomes. Cytogenetic results were available on 261 POC, fetal/pediatric samples, offspring of RBT parents. Results of PGT were available for 23 embryos obtained from two female RBT patients and a woman with multiple recurrent miscarriages attributed to a partner’s RBT.
Cytogenetic studies
Patients were referred for cytogenetic testing due to a chromosomal aberration identified in their child/fetus, RPL, infertility, or a family history of chromosome abnormality. Classical karyotype analysis was performed on peripheral blood, amniotic fluid, chorionic villi samples, or fetal tissue from POC.
Pathogenic copy number variants (CNVs) were identified by CMA, using either an oligonucleotide-based 135 K (Roche NimbleGen, Madison, WI) or a 180 K CGH + SNP (ISCA design, Agilent, Santa Clara, CA) array, as part of the clinical testing for congenital anomalies, neurodevelopmental problems, abnormal fetal ultrasound, or RPL.9 In cases suggestive of an unbalanced translocation in offspring, parental blood samples were tested by karyotype or fluorescence in situ hybridization (FISH) using subtelomere-specific probes to detect RBT.
Embryos were cultured to the blastocyst stage and biopsied on days 5/6 postfertilization. CMA was performed on amplified DNA, utilizing the 60 K GenetiSure microarray according to the manufacturer’s protocol (Agilent, Santa Clara, CA). PGT-A and PGT-SR testing were carried out using the Cytogenomics Single Cell Aberration Method software.
Statistical analysis
We performed one-tailed chi-square tests and calculated p values at the 95% confidence level to determine the differences in proportion of observed cases between male and female patients. The proportion of cytogenetic abnormalities was expected to be equal (50% each) between males and females. Variables, presented as a percentage normalized to the total number of male or female patients, were compared. Findings with p < 0.05 values were accepted as statistically significant.
RESULTS
Cytogenetic findings and reproductive outcomes
We evaluated referral reasons and reproductive outcomes in 203 couples and classified each couple into one of the following categories: couples with RPL; couples with a live-born child/(children) affected by chromosome imbalances; couples with a pregnancy with unbalanced chromosome rearrangement; couples with only progeny carrying a RBT, the same as in the parent; and couples with infertility (Fig. 1b). Further review of families with balanced progeny revealed that the majority of couples had fertility issues, including difficulty conceiving or first trimester miscarriage. Representative pedigrees are given in Fig. 2a–d. In 13 families (Fig. 2c), multiple members were found to carry a RBT without family history of an affected child or RPL. There was no difference in proportion of male and female patients in any category.
(a–d) Representative pedigrees. (a) Pedigree of a family with a translocation between chromosomes 8 and 12, carried by the father, is significant for recurrent pregnancy losses (n = 2), two affected children with the same derivative chromosome 8 (der[8]t[8;12]), and a child with a paternally inherited balanced translocation. UD undetermined karyotype. (b) Variable outcomes in a family with a cryptic three-way translocation between chromosomes 4, 18, and 20. Genetic testing has not been performed on tissues from two miscarriages. Trisomy 4 was detected by products of conception (POC) karyotyping, and trisomy 18 was reported prenatally by a cell-free DNA noninvasive prenatal testing, although confirmatory testing has not been performed. Note that offspring carry unbalanced aberrations for different chromosomes, although each is the product of missegregation of a parental balanced rearrangement. Trisomy 4 and trisomy 18 in offspring of this family are likely a product of a 3:1 segregation, misinterpreted as random aneuploidy events. (c) Three-generation pedigree showing transmission of a balanced rearrangement. There is no history of unbalanced conceptions, likely due to elimination of unbalanced gametes or lethality during the early stages of embryo or fetal development. (d) Couple with infertility attributed to a translocation between chromosomes 1 and 13, carried by the female partner. Chromosomal complements of in vitro fertilization (IVF) embryos uncovered by preimplantation testing are shown in Fig. 3. (e) A histogram depicting number of breakpoints identified on each chromosome among 203 patients with reciprocal balanced translocations (RBTs). (f) Reproductive outcomes in patients with translocations per chromosome. A graph showing a proportion of patients for each chromosome with live-born children carrying an unbalanced or a balanced chromosome complement, patients with a diagnosis of idiopathic infertility, and repeated pregnancy losses (RPL).
Parental RBTs were discovered by using two approaches: cytogenetic analysis of the parents after an unbalanced chromosome rearrangement had been detected in their offspring or POC samples by karyotype, FISH, or CMA (178 families) or karyotype analysis of patients with infertility or RPL (25 couples). Analysis of samples by CMA from offspring provided an opportunity to discover cytogenetically visible and submicroscopic aberrations, while karyotype analysis could uncover translocations of large chromosomal segments (5–10 Mb and larger). In our cohort, unbalanced submicroscopic rearrangements detected in progeny that led to the diagnosis of parental translocations were present in ~15.2% (27/178) of families. Of those, only 18% (5/27) were observed in couples with RPL, as revealed by CMA of POC samples. In the remaining 84.8% (151/178) of couples, RBT in a parent was identified after a prenatal diagnosis or the birth of a child with congenital defects or neurodevelopmental disorder.
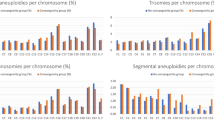
RBT involving two autosomes was detected in 196 parents. Two parents carried an X;autosome translocation, two showed translocations between the X and Y chromosomes, and two parents had a translocation involving multiple chromosomes (one with a three-way and one with a four-way RBT). In addition, one patient had two cytogenetically visible translocations, one inherited and another de novo. Eight parents (8/203) were found to carry a recurrent t(11;22)(q23.3;q11.2) translocation, while the remaining translocations had unique breakpoints. A total of 411 translocation breakpoints were observed in 203 patients. Distribution of the breakpoints by chromosome is illustrated in Fig. 2e. Chromosomes 11, 6, 4, 1, and 18 were the most commonly translocated chromosomes, detected in 18.2%, 15.3%, 14.8%, 12.3%, and 12.3% of couples, respectively. In 34.5% of the families, at least one of the translocated chromosomes was acrocentric, and imprinted chromosomes (6, 7, 11, 14, or 15) were rearranged in 48.3% of couples. Reproductive outcomes for each chromosome are shown in Fig. 2f.
Recurrent pregnancy losses in RBT patients
RPL occurred in 30.5% (62/203) of families with translocations. Among 62 families with RPL, 11 families (11/62, 17.7%) experienced the loss of an euploid fetus carrying a balanced translocation. This includes four families with paternally derived translocations and seven families with maternally inherited translocations. In 3/11 families, imprinted chromosomes 6, 11, 14, and 15 were involved in the translocation (t(6;11)(q25.1;q14.2)pat, t(15;18)(p13;p11.2)mat, t(5;14)(q13;p13)mat). Although karyotype analysis showed apparently balanced translocations in these POC samples, the presence of additional cryptic genomic alterations or regions with homozygosity cannot be excluded. In contrast to fetuses carrying balanced translocations, only two pregnancies (2/62, 3.2%) with normal karyotype were miscarried in this group.
Prevalence of aneuploid conceptions
We investigated the incidence of aneuploidy for chromosomes not involved in translocations and compared findings between males and females. The mean age of mothers was 30.5 years at the time of genetic testing, ranging from 18.9 to 42.4 years. Among miscarriages, the aneuploidy rate in the 30–32-year-old age group estimated to be ~10%.8,17 Therefore, in our set of 88 miscarried fetuses, we expected to see 9 fetuses with random aneuploidy due to maternal age, regardless if translocation was present in the mother or the father. Remarkably, we observed a total of 20 cases of aneuploidy unrelated to parental translocation (16 of 20 were among miscarriages), which is statistically higher (p < 0.024) than expected. There were 5 aneuploid fetuses among couples where a male partner carried a translocation (5/90, 5.6%) and 15 aneuploid fetuses where the mother had a translocation (15/113, 13.3%). The rate of aneuploidy in female patients was statistically different (p < 0.0027) as compared with male patients (Fig. 1c). The mean age of female patients with aneuploid fetuses was 32.6 years, while the mean age of female partners of male patients with aneuploidy was 34.4 years. Overall, the rate of aneuploidy among POC samples from a RBT male group was consistent with the aneuploidy rate in the general population, while the aneuploidy rate in female patients was significantly higher for their age group. These findings indicate that the presence of a translocation is associated with a higher risk of nondisjunction for other chromosomes during oogenesis, suggesting an interchromosomal effect in females with RBT.
Mosaic rearrangements in the parents and their offspring
Mosaic RBT was present in two individuals (2/203, 1%): a woman with 46,XX,t(11;22)(q23.3;q11.2)[6]/46,XX[14] karyotype who had a child with a nonmosaic balanced translocation, and a man with 46,XY,t(9;15)(p22;q15)[2]/46,XY[18] karyotype whose partner suffered from RPL. We also observed true chromosomal mosaicism in four POC samples: three chorionic villus samples with mosaicism for trisomy 7, trisomy 8, or trisomy 9 confined to the placenta, one with fetal mosaic trisomy 16; and one in a child carrying a balanced translocation inherited from the mother and trisomy 21 in ~10% of cells. Overall, mosaicism has been observed in ~2.7% (7/261) of offspring. Progeny of male patients demonstrated mosaicism for a tetraploid or triploid cell line along with a diploid cell line in two POC samples (2/261, 0.8%). Offspring of female patients exhibited mosaicism for aneuploidy in 5/261 (1.9%) samples, suggesting an additional risk for a postzygotic mitotic nondisjunction.
Segregation modes of balanced translocations in POC and live-born children
Balanced and unbalanced gametes can be produced in RBT patients (Fig. 1a). Although there is an equal theoretical probability for each gamete to occur, segregation of two nonhomologous chromosomes (alternate and 2:2 adjacent-1) is the most common outcome, seen in 93.2% of offspring. Other types of segregations are usually associated with extensive chromosome imbalances and are less likely to be present in POC or live-born children.
We evaluated the frequency of 2:2 adjacent-2 and 3:1 segregations. Among progeny, 3:1 segregation was observed in 15 conceptions (15/261, 5.7%), and adjacent-2 disjunction was seen in 3 samples (3/261, 1.1%). In our cohort, such malsegregations resulted in seven live-born children (7/18, 38.9%), four with Emanuel syndrome +der(22)t(11;22) (OMIM 609029), two with a supernumerary derivative chromosome 14 (+der(14)t(5;14)(q35.3;q24.2)18 and (+der(14)t(10;14)(q26;q13)) due to a 3:1 segregation, and a baby with 46,XY, + der(18)t(18;21)(q12.1;q21.2),-21 karyotype due to an adjacent-2 segregation. The remaining karyotypes were seen among POC samples. Remarkably, in 8/15 and 3/3 conceptions with 3:1 and adjacent-2 segregation, respectively, one of the translocated chromosomes was acrocentric. RBTs involving an acrocentric chromosome appear to be associated with a higher risk of a live-born child with an unbalanced chromosome complement.
Segregation modes of balanced translocations in embryos
Five euploid embryos were present among 23 tested embryos in three couples who elected to pursue IVF with PGT-A and PGT-SR. In a 35-year-old patient carrying t(6;15)(q22;q24.3), one cycle of IVF yielded two embryos, both predicted to be aneuploid. The first embryo had a double aneuploidy for monosomy 21 and trisomy 19, and the second embryo showed a high level mosaicism for monosomy X.
Nine embryos, obtained during two IVF cycles, were tested from a 30-year-old woman with RPL and two POC with paternally derived der(10)t(8;10)(q23.1;p14)pat karyotype. PGT revealed four embryos with a der(10) chromosome, two with a reciprocal der(8) abnormality, one aneuploid embryo with trisomy 11 and 21, and two (2/9, 18%) euploid embryos. All six embryos with segmental aneuploidies related to a paternal translocation were derived via 2:2 adjacent-1 segregation mode. Moreover, one out of these six embryos had a loss of 4p segment, an additional structural rearrangement.
In a 33-year-old patient with t(1;13) and infertility, 12/58 (20.7%) embryos developed to the blastocyst stage after five IVF cycles (Fig. 3). Interestingly, 8/12 embryos had segmental imbalances resulting from a malsegregation of a maternal RBT (Fig. 3h): four embryos derived from an adjacent-2 disjunction (Fig. 3a, i), one from an adjacent-1 disjunction (Fig. 3b, j), one due to a 3:1 segregation (Fig. 3c, k), one embryo due to an adjacent-1 segregation in meiosis I with a subsequent sister chromatid nondisjunction in meiosis II (Fig. 3d, l), and one embryo due to a 3:1 segregation in meiosis I with a subsequent sister chromatid nondisjunction in meiosis II (Fig. 3e, m). One embryo had chromosome imbalances unrelated to maternal translocation (Fig. 3f), and 3/58 (5.2%) embryos were euploid. Of note, one of the translocated chromosomes was acrocentric, and its malsegregation behavior was consistent with the modes observed for other patients who carry a translocation of an acrocentric chromosome.
(a–g) Whole-genome array comparative genomic hybridization (CGH) profiles showing results of preimplantation genetic testing in nine embryos. Partial ideograms for chromosomes 1 and 13 depict chromosomal aberrations in the patient with a balanced translocation (h) and in embryos with unbalanced complements (i–m). Embryo A is positive for trisomy (blue shaded area) of 1pter-q23 and monosomy (red shaded area) for 13pter-q21, consistent with the presence of a derivative chromosome 1 [der(1)] along with a normal chromosome 1 inherited from the mother (m), as shown in frame I (n = 4 embryos). Paternal complement (p), presumably normal chromosomes 1 and 13, are indicated on the right side of the image. Embryo B is positive for trisomy 1q23-qter and monosomy for 13q21-qter consistent with the presence of a derivative chromosome 13 [der(13)] along with a normal chromosome 1 inherited from the mother (m) (see j). It also had a concurrent monosomy 9. Embryo C is positive for trisomy 1q23-qter and trisomy 13pter-q21, consistent with an extra derivative chromosome 13 [der(13)], along with normal 1 and 13, transmitted from the mother (see k). Embryo D is positive for tetrasomy 1q23-qter, trisomy for 13pter-q21, and 13q21-qter monosomy, consistent with an oocyte containing two derivative chromosomes 13 along with normal 1 (l). Embryo E is positive for trisomy 1pter-q23 and 13q21-qter, and monosomy for 1q23-qter and 13pter-q21, consistent with an oocyte containing two derivative chromosomes 1 and no normal chromosome 1 or 13 (m). Embryo F is positive for monosomy 4 and trisomy for the long arm of chromosome 10. (g) A microarray plot from an euploid XX embryo.
Infertility in patients with balanced reciprocal translocations
Five males (5/90, 5.6%) and eight females (8/113, 7.1%) with RBT had a diagnosis of idiopathic infertility (Table 1). This category included childless couples, referred for karyotype analysis due to inability to conceive. In addition, two females (2/113, 1.8%) with primary ovarian insufficiency were found to carry a RBT between the X chromosome and an autosome. Chromosomes 11, 1, and 13 were the most commonly involved in a translocation among these patients. Overall, 8/13 infertile patients had a translocation involving an imprinted chromosome, and 6/13 patients carried a translocation of an acrocentric chromosome.
DISCUSSION
RBTs are well-known causes of reproductive failure, manifesting as infertility due to a depletion of germ cells, early embryonic arrest, or inability of the embryo to implant; RPL; and/or birth defects and developmental disorders in the offspring. Genetic counseling for individuals with RBT is challenging as the effect of a translocation on fertility and pregnancy outcomes depends on translocation breakpoints and other risk factors unique to each couple. In addition, postponing childbirth to 35 years of or later increases the risk of aneuploid oocytes, leading to a diminished chance for euploid pregnancy. The probabilities of different reproductive outcomes for RBT individuals are commonly based on an estimate of the likelihood of a fetus to survive with chromosomal imbalances resulting from the adjacent-1 segregation, while conceptions with other unbalanced segregations may not be viable. The risks for unbalanced offspring are calculated on the basis of empirical data19 gleaned from observations of abnormal karyotypes in POC and live births. Despite a large data set of families reported, it cannot serve as a reliable risk predictor in preconception counseling or predict the success of assisted reproductive technology (ART) for an individual couple with a RBT.
In this study, we identified additional factors predisposing to a higher risk of reproductive failure in RBT patients. Our analysis reveals a significant association between concurrent aneuploidy and female translocation status (Fig. 1c). The risk of aneuploidy in female patients is at least twofold higher (p < 0.0027) in comparison to male patients or age-matched females with normal chromosomes. Oocytes have a substantially higher aneuploidy rate than spermatozoa in general, frequently due to meiosis I nondisjunction. Meiosis I in oocytes is an already compromised process, and the abnormal quadrivalent or trivalent structure formed by translocated chromosomes may physically interfere with the segregation of other chromosomes, causing aneuploidy for a chromosome(s) uninvolved in a translocation, an interchromosomal effect (ICE).20 Our observations were made on POC from a relatively high number of natural conceptions (n = 88) lost in the second trimester of pregnancy. However, ICE may occur more frequently affecting early embryogenesis, which is impossible to evaluate from POC analysis. Also, ICE might be equally frequent in gametes from both male and female patients, although more stringent meiotic checkpoint mechanisms during spermatogenesis will lead to spermatogenic arrest and male infertility. The cause of ICE, as defined by increase in aneuploidy rate in the presence of RBT, is currently poorly understood. Animal studies suggest that disruptions in chromosome axis formation, caused by defects in checkpoint proteins or chromosome rearrangements, affect segregation of other chromosomes.21 In humans, careful analyses of the 3D spatial organization of chromosomes in the nucleus may help understand interchromosomal networks during meiosis and provide clues to ICE.
Reproductive outcomes are greatly determined by the chromosomes involved in a translocation (Fig. 2f). Our study reveals that individuals with translocation of an acrocentric chromosome are more likely to have conceptions resulting from a 3:1 segregation, causing a higher rate of miscarriage or infertility, attributable to a fetal loss in the early developmental stages (see chromosome 15 in Fig. 2f). Conversely, natural selection against embryos with chromosome imbalances implies a higher chance for a natural pregnancy being euploid (see chromosome 22, Fig. 2f). Although many unbalanced conceptions will be lost before POC can be evaluated, a high proportion of chromosomally abnormal embryos might be expected in an IVF setting, especially when the mother has a translocation.22
To establish reliable expectations and optimal treatment strategies, genetic counseling and risk calculation for couples electing ART and PGT-SR should reflect observations made of, and the success rate among, IVF patients. As seen in a patient with t(1;13), ~20% of embryos reached the blastocyst stage, and only 5% were euploid. The observed imbalances in blastocyst stage embryos provide invaluable information regarding segregation behavior of the rearranged chromosomes in human oocytes. Multiple additional factors, such as diminished ovarian reserve and patient’s age and gender, may influence the ART success in couples with RBT. It is also important to inform individuals with translocation, whether examined during prenatal or childhood testing, of cytogenetic findings when they reach a reproductive age. This will maximize patients’ chances for successful egg retrieval, storage of euploid oocytes, and reproductive decisions.
Interestingly, our analysis revealed a higher rate of pregnancy losses with a balanced translocation (17.7%) than of fetuses with apparently normal chromosomes (3.2%). The risk of a phenotypically abnormal offspring with inherited RBT is low; therefore, conceptions with balanced translocations are not expected to be miscarried. Nevertheless, the same RBTs were reported in phenotypically normal individuals and in the affected family members. Several mechanisms can explain such phenotypic variability, including the existence of cryptic imbalances or a translocation breakpoint within regulatory sequences in familial cases,23,24 which may be subject to variable penetrance and expressivity. Although rare, a gene disruption by a translocation and a pathogenic variant on another allele may result in a recessive disorder or a nonviable fetus.25 The translocation parent-of-origin effect may disturb the normal methylation pattern, affecting fetal viability. Genome sequencing (GS) and functional studies will be necessary in the future to determine the breakpoints and effects of a translocation on the imprinting and gene expression of the involved chromosome.
Increased rates of uniparental disomy (UPD) have been reported in offspring of individuals with RBT.26,27 A recessive condition or an imprinting disorder, neither of which can be detected by karyotype, may be responsible for miscarriages of fetuses with balanced translocations. This also highlights the importance of a single–nucleotide polymorphism–based chromosomal microarray (SNP-CMA) in detection of both chromosome imbalances and copy-neutral genomic abnormalities in ongoing pregnancies and POC.
Another plausible mechanism is the de novo occurrence of additional CNVs28 or structural rearrangements, as seen in our patient with two translocations. Little is known about the predisposition to structural genomic variants.29 Germline pathogenic variants in DNA repair genes may result in genome instability, as reflected by translocations, inversions, and other complex structural chromosome rearrangements, copy-number changes, single-nucleotide variations, and mosaicism in the parents or in their offspring. In some patients, inaccurate repair of DNA breaks, rather than translocation itself, may contribute to infertility,30 primary ovarian failure,31 RPL,8,32 or progeny with complex chromosome rearrangements.33 The loss of two euploid fetuses, negative for RBT, may result from de novo lethal genetic alterations or nongenetic causes. Importantly, the risk of unsuccessful pregnancies after the implantation of chromosomally normal embryos should be discussed with IVF patients undergoing PGT-SR.
In this study, mosaic RBTs were present in ~1% of parents. To date, ~35 cases with mosaic RBT were discovered.34 We also identified mosaicism in ~2.7% of the offspring of RBT patients, which included aneuploidies in conceptions from female patients and mixoploidy in offspring of male patients. This may suggest a female-specific risk for a postzygotic mitotic nondisjunction and paternal effect on zygotic segregation of parental genomes.35,36
The American Society of Reproductive Medicine recommends karyotype analysis on POC tissue for women with at least two pregnancy losses.37 Karyotype analysis has significant limitations in the detection of chromosomal rearrangements involving segments smaller than 5–10 Mb or even larger chromosomal regions with similar G-banding patterns.38 As seen in our study (Fig. 2b), karyotype analysis was useful for detection of trisomy 4 and trisomy 18 in POC and an affected infant, respectively, but was insufficient to determine the cause of adverse reproductive outcomes relevant to a paternal cryptic translocation. In addition, in ~10–40% of POC studies, fetal karyotype cannot be obtained due to culture failure or maternal cell contamination.8,12 Multiple studies have shown a significant benefit in SNP-CMA evaluation of fetal losses,11,39 including a 94–99% rate of successful results, ability to detect all chromosomal aneuploidies, triploidy, molar pregnancies, inherited and de novo submicroscopic CNVs, UPD, and complex terminal imbalances suggestive of parental RBT.10,12
In our cohort, 15% of couples had RBT below the detection resolution of classical karyotype. In these families, parental rearrangements were diagnosed after the birth of an affected child or a miscarriage, and a positive CMA. Importantly, chromosomal imbalances smaller than 10 Mb will be missed by current PGT-A and PGT-SR technologies and are undetectable by cell-free DNA noninvasive prenatal testing. The diagnosis of a balanced chromosome rearrangement in parents who have no apparent abnormal phenotype is strictly dependent on accurate genetic analysis of their offspring. Multiple professional societies recommend CMA as a first-tier test in prenatal and postnatal diagnosis due to a much higher diagnostic yield, technical advances, and superior sensitivity in diagnosis of cryptic chromosome imbalances.40 The application of SNP-CMA in POC evaluation as a standard of care will lead to an accurate and early diagnosis, reduction of emotional impact associated with RPL, and significant improvement in the clinical management of infertility and future pregnancies.
Recent studies on patients with RPL indicated that ~12% of couples have chromosomal abnormalities, ~40% of which are undetectable by karyotype.7 The incidence of cryptic balanced chromosomal rearrangements in patients with infertility is known; thus, undiagnosed couples will have a higher risk of implantation of embryos with segmental imbalances that have lower potential for implantation or normal fetal development. The identification of a parental balanced rearrangement is imperative to provide appropriate genetic counseling or explore IVF options to have a healthy child. When genetic testing of POC tissues is not an option, detection of parental rearrangements by GS may improve the odds of IVF and selection of chromosomally normal embryos for implantation.
Present analysis is limited by a small number of samples for each category studied. Our findings warrant further research to develop a genome-based risk assessment tool that will assist in interpretation of genomic structural rearrangement based on the current knowledge of dosage-sensitive genes, genomic structure, noncoding regulatory regions, epigenomic features, chromosome-specific behavior in meiosis, and clinical observations.
Data availability
All raw data are available from the corresponding author.
References
Halgren, C. et al. Risks and recommendations in prenatally detected de novo balanced chromosomal rearrangements from assessment of long-term outcomes. Am. J. Hum. Genet. 102, 1090–1103 (2018).
Van Dyke, D. L., Weiss, L., Roberson, J. R. & Babu, V. R. The frequency and mutation rate of balanced autosomal rearrangements in man estimated from prenatal genetic studies for advanced maternal age. Am. J. Hum. Genet. 35, 301–308 (1983).
Morin, S. J., Eccles, J., Iturriaga, A. & Zimmerman, R. S. Translocations, inversions and other chromosome rearrangements. Fertil. Steril. 107, 19–26 (2017).
Elghezal, H., Hidar, S., Braham, R., Denguezli, W., Ajina, M. & Saâd, A. Chromosome abnormalities in one thousand infertile males with nonobstructive sperm disorders. Fertil. Steril. 86, 1792–1795 (2006).
Yatsenko, A. N. et al. Comprehensive 5-year study of cytogenetic aberrations in 668 infertile men. J. Urol. 183, 1636–1642 (2010).
Priya, P. K., Mishra, V. V., Roy, P. & Patel, H. A study on balanced chromosomal translocations in couples with recurrent pregnancy loss. J. Hum. Reprod. Sci. 11, 337–342 (2018).
Dong, Z. et al. Genome sequencing explores complexity of chromosomal abnormalities in recurrent miscarriage. Am. J. Hum. Genet. 105, 1102–1111 (2019).
Yatsenko, S. A. et al. Cytogenetic signatures of recurrent pregnancy losses. Prenat. Diagn. 41, 70–78 (2021).
Yatsenko, S. A. et al. Application of chromosomal microarray in the evaluation of abnormal prenatal findings. Clin. Genet. 84, 47–54 (2013).
Levy, B. et al. Genomic imbalance in products of conception: single-nucleotide polymorphism chromosomal microarray analysis. Obstet. Gynecol. 124, 202–209 (2014).
Peters, D. G., Yatsenko, S. A., Surti, U. & Rajkovic, A. Recent advances of genomic testing in perinatal medicine. Semin. Perinatol. 39, 44–54 (2015).
Sahoo, T. et al. Comprehensive genetic analysis of pregnancy loss by chromosomal microarrays: outcomes, benefits, and challenges. Genet. Med. 19, 83–89 (2017).
Hay, S. B. et al. ACOG and SMFM guidelines for prenatal diagnosis: is karyotyping really sufficient? Prenat. Diagn. 38, 184–189 (2018).
Wilch, E. S. & Morton, C. C. Historical and clinical perspectives on chromosomal translocations. Adv. Exp. Med. Biol. 1044, 1–14 (2018).
Zhang, W. et al. Clinical application of next-generation sequencing in preimplantation genetic diagnosis cycles for Robertsonian and reciprocal translocations. J. Assist. Reprod. Genet. 33, 899–906 (2016).
Xie, Y. et al. Preliminary analysis of numerical chromosome abnormalities in reciprocal and Robertsonian translocation preimplantation genetic diagnosis cases with 24-chromosomal analysis with an aCGH/SNP microarray. J. Assist. Reprod. Genet. 35, 177–186 (2018).
Grande, M. et al. The effect of maternal age on chromosomal anomaly rate and spectrum in recurrent miscarriage. Hum. Reprod. 27, 3109–3117 (2012).
Bregand-White, J., Saller, D. N., Clemens, M., Surti, U., Yatsenko, S. A. & Rajkovic, A. Genotype-phenotype correlation and pregnancy outcomes of partial trisomy 14q: a systematic review. Am. J. Med. Genet. A. 170, 2365–2371 (2016).
Stengel-Rutkowksi, S., Stene, S. & Gallano, P. Risk estimates in balanced parental reciprocal translocations. Monographie des Annales de Genetique (Expansion Scientifique Francaise, Paris 1988).
Mateu-Brull, E. et al. Interchromosomal effect in carriers of translocations and inversions assessed by preimplantation genetic testing for structural rearrangements (PGT-SR). J. Assist. Reprod. Genet. 36, 2547–2555 (2019).
Miller, D. E. The interchromosomal effect: different meanings for different organisms. Genetics. 216, 621–631 (2020).
Zhang, L. et al. Interaction of acrocentric chromosome involved in translocation and sex of the carrier influences the proportion of alternate segregation in autosomal reciprocal translocations. Hum. Reprod. 34, 380–387 (2019).
Ordulu, Z. et al. Structural chromosomal rearrangements require nucleotide-level resolution: lessons from next-generation sequencing in prenatal diagnosis. Am. J. Hum. Genet. 99, 1015–1033 (2016).
Schluth-Bolard, C. et al. Cryptic genomic imbalances in de novo and inherited apparently balanced chromosomal rearrangements: array CGH study of 47 unrelated cases. Eur. J. Med. Genet. 52, 291–296 (2009).
Hearn, T. et al. Mutation of ALMS1, a large gene with a tandem repeat encoding 47 amino acids, causes Alström syndrome. Nat. Genet. 31, 79–83 (2002).
Liehr, T. Cytogenetic contribution to uniparental disomy (UPD). Mol. Cytogenet. 3, 8 (2010).
Behnecke, A., Hinderhofer, K., Jauch, A., Janssen, J. W. G. & Moog, U. Silver-Russell syndrome due to maternal uniparental disomy 7 and a familial reciprocal translocation t(7;13). Clin. Genet. 82, 494–498 (2012).
Van Binsbergen, E., Hochstenbach, R., Giltay, J. & Swinkels, M. Unstable transmission of a familial complex chromosome rearrangement. Am. J. Med. Genet. A. 158A, 2888–2893 (2012).
Javadekar, S. M. & Raghavan, S. C. Snaps and mends: DNA breaks and chromosomal translocations. FEBS J. 282, 2627–2645 (2015).
Cheung, S., Parrella, A., Rosenwaks, Z. & Palermo, G. D. Genetic and epigenetic profiling of the infertile male. PLoS One. 14, e0214275 (2019).
Katari, S. et al. Chromosomal instability in women with primary ovarian insufficiency. Hum. Reprod. 33, 531–538 (2018).
Zhang, W. et al. KHDC3L mutation causes recurrent pregnancy loss by inducing genomic instability of human early embryonic cells. PLoS Biol. 17, e3000468 (2019).
Wood-Trageser, M. A. et al. MCM9 mutations are associated with ovarian failure, short stature, and chromosomal instability. Am. J. Hum. Genet. 95, 754–762 (2014).
Garzo, M. et al. Ten new cases of balanced reciprocal translocation mosaicism (BRTM): reproductive implications, frequency and mechanism. Eur. J. Med. Genet. 63, 103639 (2020).
Carson, J. C. et al. Diploid/triploid mixoploidy: a consequence of asymmetric zygotic segregation of parental genomes. Am. J. Med. Genet. A. 176, 2720–2732 (2018).
Destouni, A. et al. Zygotes segregate entire parental genomes in distinct blastomere lineages causing cleavage-stage chimerism and mixoploidy. Genome Res. 26, 567–578 (2016).
Practice Committee of the American Society for Reproductive Medicine. Evaluation and treatment of recurrent pregnancy loss: a committee opinion. Fertil. Steril. 98, 1103–1111 (2012).
Marinescu, P. S., Saller, D. N., Parks, W. T., Yatsenko, S. A. & Rajkovic, A. Prenatal diagnosis of trisomy 6q25.3-qter and monosomy 10q26.12-qter by array CGH in a fetus with an apparently normal karyotype. Clin. Case Rep. 3, 92–95 (2015).
Popescu, F., Jaslow, C. R. & Kutteh, W. H. Recurrent pregnancy loss evaluation combined with 24-chromosome microarray of miscarriage tissue provides a probable or definite cause of pregnancy loss in over 90% of patients. Hum. Reprod. 33, 579–587 (2018).
Committee on Genetics and the Society for Maternal-Fetal Medicine. Committee opinion no. 682: microarrays and next-generation sequencing technology: the use of advanced genetic diagnostic tools in obstetrics and gynecology. Obstet. Gynecol. 128, e262–e268 (2016).
Acknowledgements
We are grateful to the staff of the Pittsburgh Cytogenetics Laboratory for the technical assistance in cytogenetics studies.
Author information
Authors and Affiliations
Contributions
Conceptualization: A.R., S.A.Y. Data curation: E.S., S.D., L.W., R.C., A.V., S.A.Y. Formal analysis: J.H., U.S., M.B., S.A.Y. Investigation: A.V., E.S., M.C., S.D., L.W., R.C., S.K., J.S., S.A.Y. Methodology: A.V., D.N.S., A.R.; S.A.Y. Supervision: S.A.Y. Writing—original draft: A.V., S.A.Y. Writing—review & editing: J.H., U.S., M.B., E.S., M.C., S.D., L.W., R.C., S.K., J.S., D.N.S., A.R., S.A.Y.
Corresponding author
Ethics declarations
Ethics declaration
The study was approved by the University of Pittsburgh IRB (STUDY20060192; STUDY18120172; STUDY20050099). The detailed PGT results of an individual patient are included as an example. The signed consent form was obtained from that patient.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Verdoni, A., Hu, J., Surti, U. et al. Reproductive outcomes in individuals with chromosomal reciprocal translocations. Genet Med 23, 1753–1760 (2021). https://doi.org/10.1038/s41436-021-01195-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-021-01195-w
This article is cited by
-
Low androgen signaling rescues genome integrity with innate immune response by reducing fertility in humans
Cell Death & Disease (2024)
-
What proportion of couples with a history of recurrent pregnancy loss and with a balanced rearrangement in one parent can potentially be identified through cell-free DNA genotyping?
Molecular Cytogenetics (2023)
-
Are ovarian responses and the number of transferable embryos different in females and partners of male balanced translocation carriers?
Journal of Assisted Reproduction and Genetics (2022)