Abstract
Purpose
The increasing use of electronic health records (EHRs) and biobanks offers unique opportunities to study Mendelian diseases. We described a novel approach to summarize clinical manifestations from patient EHRs into phenotypic evidence for cystic fibrosis (CF) with potential to alert unrecognized patients of the disease.
Methods
We estimated genetically predicted expression (GReX) of cystic fibrosis transmembrane conductance regulator (CFTR) and tested for association with clinical diagnoses in the Vanderbilt University biobank (N = 9142 persons of European descent with 71 cases of CF). The top associated EHR phenotypes were assessed in combination as a phenotype risk score (PheRS) for discriminating CF case status in an additional 2.8 million patients from Vanderbilt University Medical Center (VUMC) and 125,305 adult patients including 25,314 CF cases from MarketScan, an independent external cohort.
Results
GReX of CFTR was associated with EHR phenotypes consistent with CF. PheRS constructed using the EHR phenotypes and weights discovered by the genetic associations improved discriminative power for CF over the initially proposed PheRS in both VUMC and MarketScan.
Conclusion
Our study demonstrates the power of EHRs for clinical description of CF and the benefits of using a genetics-informed weighing scheme in construction of a phenotype risk score. This research may find broad applications for phenomic studies of Mendelian disease genes.
Similar content being viewed by others
INTRODUCTION
Cystic fibrosis (CF) is a recessive Mendelian disease caused by a spectrum of pathogenic variants in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. As one of the most common Mendelian diseases, CF continues to pose challenges due to the highly variable clinical manifestations displayed among CF patients.1 Part of the variability reflects the spectrum of pathogenic variants in the CFTR gene, which differ in impact on disease onset, severity, and treatment.2,3,4 However, the phenotypic variation in CF cannot be explained by the CFTR coding variants alone. A variety of studies have identified variants in other regions of the genome that impact the CF phenotypic variability.5,6,7 It remains to be seen whether regulatory variants modulating the expression of CFTR might add to the phenotypic variability. Presumably, regulatory variation of Mendelian genes would cause milder phenotypes; in support of this, genome-wide association studies (GWAS) of common diseases have revealed overrepresentation of Mendelian genes among the identified risk loci.8 On the other hand, regulatory variants can also act to modify (reduce) the deleteriousness of coding variants, as shown in cancers and autism.9
In this study, we proposed to interrogate the phenotypic consequences of regulatory variants of CFTR. The aggregate effects of multiple regulatory variants in a gene were determined by using genotypes to impute genetically regulated expression (GReX) from reference resources such as the Genotype–Tissue Expression (GTEx) database.10,11 Clinical outcomes of predicted expression of CFTR were examined through a phenome-wide association study (PheWAS,12 an unbiased test of association of a genotype with a range of clinical diagnoses) in BioVU, an academic medical center–based biobank with genotypes linked to electronic health records (EHRs).13 Moreover, we evaluated in an independent data set containing EHRs from 2.8 million patients how well the identified EHR phenotypes in aggregate predicted clinically diagnosed CF.
MATERIALS AND METHODS
Data sources
Data were obtained from Synthetic Derivative (SD), the de-identified clinical data warehouse at Vanderbilt University Medical Center (VUMC), and BioVU, the VUMC biobank that contains >250,000 DNA samples. All the data were de-identified and our study was classified as “nonhuman subjects” research by the VUMC Institutional Review Board in accordance with the provisions of Title 45, Code of Federal Regulations, part 46. The genotype data set contains genome-wide genotype data from 9142 BioVU participants of European ancestry. The genotypes were imputed and phased into Human Haplotype Reference Consortium reference panel (version r.1.1)14,15 using IMPUTE2.16 Samples of European ancestry were extracted for analysis based on ancestry principal component analysis (PCA). Our second data set contains phenotype-only data from 2.8 million patients of SD (excluding the 9142 BioVU participants).
Imputing CFTR expressions from genotypes
Expression imputation models were previously trained on the GTEx reference panel (version 2015).17 GReX of CFTR in each tissue was calculated as a weighted sum of the composite alleles in the prediction model. Of the 20 tissue-specific prediction models available for CFTR, we focused on the models with modest prediction performance (i.e., correlation of at least 0.1 between predicted and measured expression), and applied the models to the individual-level genotypes of BioVU samples to calculate GReX. We further used phasing information of the genotype data to impute GReX at haplotype-level (hGReX) for tissue “brain hypothalamus.”
PheWAS
PheWAS of the GReX of CFTR was performed in each tissue separately via logistic regression, adjusting for age, gender, three principal components of ancestry, and arrays/batches. The binary phenotypes (“phecodes”) were derived from billing codes of EHRs as described previously12,18 with the use of the PheWAS package.19 Each phecode has defined case, control, and exclusion criteria and we required two codes on different visit days to instantiate a case for each phecode. Only phecodes with at least 20 cases were included in analysis. Effect sizes were reported by the beta estimates from the regression.
LD-proxy of DF508
DF508 (CFTR p.Phe508del) is a three–base pair deletion (rs113993960, 7:117199645-ATCT-A) on the 508th codon of the CFTR gene. Since DF508 was not directly genotyped in initial genotyping arrays, we used the linkage disequilibrium (LD)-proxy allele to tag it (rs111309367_T, r2 = 0.4, D’ = 1). While D’ = 1, DF508 is less common than this proxy allele; we have P(proxy=1 | DF508 = 1)=1 and P(DF508 = 0 | proxy = 0) = 1. The latter condition indicates 100% specificity of the proxy allele (i.e., noncarriers of DF508proxy are also noncarriers of DF508). The former condition can be used to simply the calculation of sensitivity (of the proxy allele to tag DF508) into a ratio of two allele frequencies (AF):
With 1 in 2500 newborns with an incidence of CF being of European descent20 and DF508 being present on 69–76% of cystic fibrosis chromosomes in North American CF patients,21,22 we estimated that the allele frequency (AF) of DF508 in population of European ancestry is approximately 1.67%. This is derived as follows: proption_of_CF_patients_with_DF508 = P2 + 2P(0.5p) = 2P2, and the proportion_of_CF_patients_with_DF508 also equals \(\frac{1}{{2500}}\left( {0.7} \right)\). So \(2p^2 = \frac{1}{{2500}}\left( {0.7} \right)\), thus p2 = sqrt(1/2500*0.7) = 1.67%. Given an AF of 2% for the proxy allele in (non-Finnish) European descent (gnomAD [gnomad.broadinstitute.org]; haploreg4 [pubs.broadinstitute.org/mammals/haploreg/haploreg.php]), the sensitivity was estimated ~80% (=1.67%/2%). This implies that carriers of DF508proxy are not necessarily also carriers of DF508—a portion of the homozygotes (heterozygotes) of DF508proxy are actually heterozygous (non)carriers of DF508. We denote this proxy allele as DF508proxy.
GReX of CFTR between carriers and noncarriers of CF-pathogenic alleles
In addition to DF508, we interrogated additional CF-pathogenic alleles (according to ClinVar [version 2017]) that were covered by our genotype data, collectively denoted as “other” CF alleles. Heterozygous carriers of these “other” CF alleles were carefully determined as carriers of one of these “other” CF-pathogenic alleles who neither carry (1) DF508proxy nor (2) a diagnosis of CF. Condition 2 was to exclude potential compound heterozygotes who carry CF-pathogenic alleles uncovered by our genotyping arrays. We tested for difference in hGReX between heterozygous carriers and noncarriers of (1) DF508 and (2) “other” CF-pathogenic alleles using nonparametric Wilcoxon signed-rank test.
Measured expression of CFTR in relation to DF508
We examined the measured expression of CFTR stratified by the dosage of DF508 using the expression data (RNA-seq) and matched genome sequencing data from GTEx (V8 release). We focused on tissues with an averaged expression level of CFTR above a threshold (transcript per million [TPM] ≥0.01 in GTEx v7). Gene expressions in each tissue were processed according to ref. 23 including steps of quantile normalization, adjustment for covariates (gender, platform, first five principal components [PCs], and probabilistic estimation of expression residual [PEER] factors to remove hidden batch effects and other confounders in the expression data), and regression of the expression residuals against the dosage of DF508.
Phenotype risk score construction and performance evaluation
In a data set (“validation set”) that contains EHRs from 2.8 million patients (excluding the 9142 participants of the discovery set) from the SD of VUMC, we constructed and evaluated three phenotype risk scores (PheRSs). The EHR phenotypes and weights used to construct each PheRS (PheRSmapping, PheRSassoc, and PheRShybrid) are shown in Supplementary Table S2. The weights for PheRSmapping were extracted from the original paper12 based on disease prevalence estimated in VU individuals of European ancestry. Since only the relative values matter for the weights, we normalized the weights to have the sum equal to 1. Both the weights of PheRSassoc and PheRShybrid were beta (effect size) values from GReX–phenotype associations and normalized to sum up to 1.
The performance of the PheRSs for differentiating CF cases (defined as having the CF diagnosis code in EHRs) from controls was assessed via logistic regression to obtain the probability of the disease occurrence. Because of the highly unbalanced data (~0.1% of CF cases), we calculated the average precision rate (i.e., the area under precision recall curve) to measure model performance. Each time, 150,000 patients were randomly selected from the validation set, and the average precision was evaluated for both methods (PheRSassoc vs. PheRSmapping). We repeated this process ten times and compared the performance.
Evaluation of PheRSs in MarketScan
The MarketScan databases, owned by IBM Watson Health, are a suite of administrative claims-based databases that comprise inpatient and outpatient claims, medical procedure claims, prescription claims, clinical utilization records, and health-care expenditures. These data are collected from employers, managed care organizations, health plan providers, and state Medicaid agencies. The covered patient population includes more affluent, privately insured segments of US society.24,25 The MarketScan databases describe over half of the US population in terms of comprehensive and high-quality coding of diagnoses, procedures, and drug prescriptions. There have been more than 900 peer-reviewed publications since the launch of these databases in 1995, and this number has increased even more rapidly in recent years.26,27
To further evaluate the proposed PheRSs in this study, we used one of the MarketScan databases—the MarketScan Commercial Claims and Encounters database.28 This commercial database contains medical claims, outpatient prescription drug claims, and person-level enrollment information. We identified 25,314 CF cases whose first CF diagnosis appearing in the database was at age of 30 years or older and randomly selected 99,991 non-CF controls who are age- and gender-matched to the CF cases, of a total of 151 million unique individuals enrolled in the database during the years 2003–2013.
RESULTS
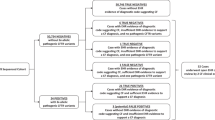
The workflow of the study is described in Fig. 1.
EHR phenotypes associated with genetically determined expression of CFTR
Using the expression imputation models previously trained on the GTEx reference panel,17 we estimated tissue-specific GReX of CFTR in ten tissues with modest prediction performance (R2 of at least 0.01; Supplementary Table S1). Phenome-wide scan of the GReX of CFTR was performed in BioVU participants of European ancestry (n = 9142). In brain hypothalamus, the GReX was associated with clinically diagnosed cystic fibrosis (P = 2.3 × 10−39). Other top-ranked associations reflect clinical symptoms in respiratory, endocrine and metabolic, and gastrointestinal systems (Table 1). These phenotypes capture key classic features of CF, such as pseudomonal pneumonia (P = 1.6 × 10−26), MRSA pneumonia (i.e., methicillin susceptible pneumonia due to Staphylococcus aureus, P = 1.3 × 10−20), bronchopneumonia and lung abscess (P = 8.4 × 10−14), and bacterial pneumonia (P = 6.2 × 10−12) for respiratory manifestations; disease of pancreas (P = 2.1 × 10−17) and secondary diabetes (P = 5.0 × 10−9) for endocrine and metabolic manifestations; and nutritional marasmus (low weight in infant/child) (P = 1.1 × 10−8), intestinal malabsorption (nonceliac) (P = 1.1 × 10−7), severe protein calorie malnutrition (P = 0.0001), failure to thrive in childhood (P = 1.2 × 10−6), and lack of normal physiological development (P = 6.7 × 10−6) for gastrointestinal manifestations (Table 1). The top associations also include rarer phenotypes such as bronchiectasis (P = 4.9 × 10−19), hemoptysis (coughing up blood or blood-stained mucus, P = 1.6 × 10−5), as well as common phenotypes including nasal polyps (P = 2.6 × 10−5), abnormal sputum (P = 2.6 × 10−5), and chronic sinusitis (P = 8.5 × 10−5). These milder symptoms are consistent with previously reported symptoms in CF cases with adult onset.29,30,31,32
We denoted the top 20 associated EHR phenotypes (excluding CF diagnosis) detected in hypothalamus collectively as the CF-phenome (Table 1). Notably, the direction of association was concordantly negative for these top associations (i.e., risk of symptoms was inversely related to the GReX level of CFTR in hypothalamus). Similar phenome associations (but less comprehensive) were also detected in two other tissues (brain hippocampus, heart left ventricle) (Supplementary Table S2).
GReX of CFTR captures underlying CF coding alleles
Given that the GReX associations captured CF and many of its clinical manifestations, we asked whether the GReX reflects a genuine effect of regulatory variants independent of coding variants, or mainly captures the coding variants in CFTR due to LD. We first conditioned our analysis on DF508, the most common CF-pathogenic variants in people of European descent. Since DF508 was not directly genotyped in our genotyping arrays, we used the LD-proxy allele (rs111309367, r2 = 0.4, D’ = 1) that tags DF508 with 100% specificity and ~80% sensitivity (see “Materials and Methods”). We denoted this proxy allele as DF508proxy.
After conditioning on the dosage of DF508proxy, the association of GReX of CFTR (in hypothalamus) with the CF-phenome attenuated sharply (Table 1). Indeed, GReX of CFTR was correlated with the dosage of DF508proxy, showing a dosage-dependent trend with respect to DF508proxy (Fig. 2a). None of the individual single-nucleotide polymorphisms (SNPs) that comprise the GReX in hypothalamus are, however, in strong LD with DF508proxy (r2 < 0.2) (Supplementary Fig. S1). We hypothesized that it is the combination of the noncoding alleles on haplotypes that effectively capture DF508proxy. To investigate this, we decomposed the GReX into the sum of two haplotype-level predicted gene expressions (hGReX) assuming an additive model (see “Materials and Methods”). With phased genotype data, we observed that in heterozygotes of DF508proxy (n = 414, excluding CF patients), the haplotype carrying DF508proxy almost exclusively (98.7%) had lower hGReX than the other (wild-type) haplotype (Wilcoxon signed-rank test P < 2.2 × 10−16; Fig. 2b).
a GReX stratified by the dosage of DF508proxy. b Haplotype-level GReX (hGReX) in heterozygous carriers of DF508proxy who were not diagnosed as cystic fibrosis (CF) (n = 414). Each heterozygote is represented by a pair of dots, with red referring to the haplotype carrying DF508proxy and black the other wild-type haplotype.
We then checked whether the expression reduction was also seen in haplotypes carrying CF alleles other than DF508. There are 16 additional CF alleles (according to ClinVar [version 2017]) covered either by our direct genotyping or genotype imputation (Supplementary Table S3). With the allele frequency ranging from 0.001% to 0.2% in BioVU samples, we observed that individuals either carry zero or a single CF allele. Of the carriers (n = 121), a few were positive for DF508proxy (n = 14) or CF case status (n = 4); after exclusion of these individuals, we obtained 103 heterozygous carriers for one of these 16 CF alleles who were without diagnosis of CF (Fig. 3a). In these heterozygotes, the haplotype carrying a CF allele on average had lower hGReX than the wild-type haplotype (Wilcoxon signed-rank test; P < 4.7 × 10−12; Fig. 3b), similar to the observation of DF508proxy. In contrast, the load of intronic variants was not correlated with the level of hGReX (P = 0.8).
a Sample distribution by genotype and CF case status. Case is defined by the presence of CF diagnosis code in electronic health records (EHRs). “Other” indicates 1 of 16 CF-pathogenic alleles that are also covered by our genotype data. b hGReX of haplotypes harboring DF508proxy (n = 414), of haplotypes harboring one of “other” CF-pathogenic alleles (n = 103), of wild-type haplotypes from the same carriers (n = 103), and of haplotypes from the remaining noncarriers. Het heterozygous, homo homozygous.
Measured expression of CFTR in carriers of DF508
Using the genome sequencing from more than 800 individuals of GTEx database (release V8), we examined the measured gene expression in relation to DF508 in three tissues (hypothalamus, hippocampus, and heart left ventricle) where CF-phenome was detected. The number of carriers of DF508 is small in all three tissues (6–8 heterozygous carriers). In brain hippocampus we detected expression reduction of CFTR in carriers of DF508 (Wilcoxon rank sum test; P = 0.006), while no difference was detected in the other two tissues (Supplementary Fig. S2), likely due to the better correlation between GReX and the actual expressions in hippocampus (r2 = 0.074) than in hypothalamus (r2 = 0.011) or heart left ventricle hypothalamus (r2 = 0.025).
Scoring individuals based on EHR phenotypes identified by GReX associations
Since our GReX-associated phenotypes are consistent with clinical features of CF, we assessed whether these EHR phenotypes can be combined to construct a phenotype score to express how close an individual’s EHR phenotypes are to clinically diagnosed CF. Earlier attempts built a phenotype risk score for CF by mapping clinical description of Mendelian diseases to EHR phenotypes and then aggregating the relevant EHR phenotypes into a weighted sum with the weights determined by the inverse prevalence of the phenotypes in EHRs.33 We denoted this score as PheRSmapping (for the composite EHR phenotypes and weights, see Supplementary Table S4).
Here, we constructed an alternative PheRS for CF: we combined the GReX-discovered CF-phenome (20 phecodes, excluding CF diagnosis, phecode 499) using weights informed by the effect size estimates from the GReX–phenotype associations (see “Materials and Methods”; Supplementary Table S5). This phenotype risk score, denoted as PheRSassoc, scored CF patients higher than controls (Wilcoxon rank sum test; P < 2.2 × 10−16) in samples independent of those used discovering the CF-phenome (N = 31,537 European-ancestry samples [EUs], with 131 CF cases), validating PheRSassoc as a phenotype score for CF (“Materials and Methods”).
Next, we compared the performance of PheRSassoc with PheRSmapping using de-identified EHRs from 2.8 million patients from VUMC (~0.1% were diagnosed as CF cases), independent of the discovery data set. The precision and recall rates were compared side by side for the scores for ten iterations, and each time a random sample of 150,000 individuals (EHRs) was selected from the total pool (“Materials and Methods”). For each of the ten data sets, the average precision rate (i.e., area under the precision recall curve) of PheRSassoc is better than PheRSmapping, ranging from 20% to 36% for the former and from 3% to 12% for the latter (Fig. 4; Supplementary Fig. 3; Supplementary Table S6). Consistently, the precision and recall of predicted high-risk patients (defined as the top 100 high-scoring individuals) of PheRSassoc were better than PheRSmapping across all ten iterations (Supplementary Table S7).
a Phecodes and weights used to construct PheRSassoc, PheRSmapping, and PheRShybrid. Orange and blue indicate phecodes specific to PheRSassoc and PheRSmapping, respectively; gray indicates shared phecodes. PheRShybrid by design has the same constitutive phecodes as PheRSmapping with weights derived from genetically regulated expression (GReX) associations (NA indicates weights not available due to logistic regression not performed for case number <20). b Performance (area under precision recall curves) across ten iterations are shown, each with 150,000 patients randomly sampled from a data set containing de-identified electronic health records (EHRs) from 2.8 million patients that do not overlap the discovery data set. MRSA methicillin-resistant Staphylococcus aureus.
Since the number of phecodes used in constructing each PheRS is similar (21 phecodes in PheRSmapping vs. 20 phecodes in PheRSassoc, and 4 shared phecodes between the two scores), we hypothesized that the different weighting schemes may have contributed to the performance difference. To test this, for the PheRSmapping, we kept the constitutive phenotypes unchanged but replaced the original weights with the weights derived from the GReX–phenotype associations detected in hypothalamus (Fig. 4a); the performance of the resulting PheRS (denoted as PheRShybrid) almost tripled in the precision recall rate, ranging from 11% to 23% (Fig. 4b). This indicates that the genetics-informed weights substantially outperformed the prevalence-based weights for predicting case status of CF. In addition, the constitutive codes of PheRSassoc generally have better discriminative power for CF than the codes of PheRSmapping, as indicated by the logistic regression of each code against CF status (affected vs. unaffected) that generated larger odds ratios of the codes of PheRSassoc (Supplementary Table S8).
Applying PheRSassoc to the case presentation of a 47-year-old woman who was diagnosed with CF in adulthood,29 the woman ranked in the 99.9th percentile for CF among 2.8 million VUMC patients (Supplementary Table S9), suggesting the potential of our PheRSassoc to effectively alert possible CF cases with adult onset. As a comparison, the PheRSmapping scoring ranked the same woman as in the 98th percentile for CF.34 Case presentations of the woman fit 9/20 phenotype components of PheRSassoc, including sinusitis, cough, and abnormal sputum, which were not part of the components of PheRSmapping.
We further evaluated the PheRSs in MarketScan, an independent database that contains national-level EHRs from nearly half of the US population28 (“Materials and Methods”). After mapping the International Statistical Classification of Diseases and Related Health Problems (ICD) codes to phecodes, we applied the scoring algorithms to adults aged 30 years or older (“Materials and Methods”). We found that (1) PheRSassoc can distinguish CF cases from non-CF controls (one-sided Wilcoxon rank sum test, P < 3.2E-249) and (2) PheRSassoc consistently performed better than PheRSmapping (Supplementary Table S10).
DISCUSSION
In this work, we demonstrate that the genetically regulated expression of a gene (CFTR) causing a Mendelian disease can be used as a genetic instrument to identify EHR phenotypes consistent with the Mendelian disease (CF). The associated EHR phenotypes can be combined effectively into a PheRS to summarize the evidence of phenotype overlap with CF. The novel weighting scheme guided by the phenotypic associations enhanced the accuracy of PheRS for predicting CF case status. Given that primary care physicians are estimated to encounter 2–3 cases of CF over the course of their clinical practice,35 it is important to recognize CF cases in adults whose clinical manifestations tend to deviate from those with early onset. The potential of our PheRS to identify possible CF with onset in adulthood points to the clinical utility of this study. With continuous expansion of EHRs and biobanks, our phenotype risk score will continue to evolve, and may eventually facilitate earlier identification of adult onset of CF.
It has been established that specific cells in lung, ionocytes, a minority cell type in lung, express CFTR proteins leading to the canonical lung phenotypes associated with CF.36,37 It is therefore not surprising that we did not detect CF-phenome associations from lung that contains bulk expressions of various cell types. In human brain, hypothalamus is the first site of brain discovered for CFTR expression,38 and only neurons were found to express CFTR proteins.39,40 Lineage relationship traces neurons back to intermediate neuronal progenitors (a form of basal progenitor)41 and basal progenitors are known to also generate ionocytes.39,40 We speculate that brain hypothalamus includes a cell type that shares a developmental lineage with ionocytes in lung and that similar such cell types are present in the other tissues for which we see strong associations to CFTR phenotype. This implies that the cell types expressing CFTR in brain hypothalamus and the other tissues we observed to show strong associations to CF phenotypes also have higher proportions of cells with a potentially related developmental ontology to the lung ionocytes implicated in CF.
We presented a de novo approach that simultaneously identifies the components required for a phenotype risk score: clinical phenotypes and their corresponding weights. The constitutive codes of PheRSassoc in general have a better discriminative power for CF than the codes of PheRSmapping. The weights, which are proportional to the effect sizes, reflect the relative importance of each component EHR phenotype on CF diagnosis (bi / bCF) as they were measured by a common genetic instrument (genetically determined expression). The genetics-informed weights perform better than the prevalence-based weights as the latter do not capture such relational importance to EHR-based CF diagnosis.
Another contributing factor to the improved performance of our de novo approach is that our approach exploits the rich and detailed EHR phenotypes. For example, pneumonia is included in the clinical description of CF, and was mapped to EHR “pneumonia” (phecode 480). Our association analysis revealed additional forms of pneumonia, such as “bacterial pneumonia” (phecode 480.1), “pseudomonal pneumonia” (phecode 480.12), and “methicillin susceptible pneumonia due to Staphylococcus aureus” (phecode 480.13). These pneumonia terms were all more strongly associated with GReX of CFTR (P < 7 × 10−11) than the general term “pneumonia” (P = 0.02). This indicates that our de novo approach circumvents some of the difficulties in mapping clinical description terms to EHR phenotypes, which are structured hierarchically.
Our results do not support a causal role of predicted expression of CFTR on CF phenotypes. The lowest predicted expression was also seen in controls; however, in CF patients, there was an overrepresentation of the low levels of GReX. Additionally, when we repeated the analysis by excluding the 71 individuals with CF diagnosis (the remaining 9071 patients), all the association signals regarding the CF-phenome disappeared (data not shown), suggesting the predicted expression of CFTR is unlikely to be an independent or significant contributor to CF phenotypes, at least at these sample sizes. The observed coupling of CF variants with expression-reducing alleles is consistent with the hypothesis that natural selection favors haplotypes whose composite regulatory alleles reduce the functional impact of the deleterious variants.9 In line with this explanation, the haplotypes harboring a severe CF allele such as DF508 demonstrated a lower GReX than the haplotypes harboring a less severe CF allele (Fig. 3b). In this regard, since the level of GReX coevolves with the deleteriousness of total underlying CF alleles due to natural selection, the effect size estimates based on GReX in fact capture the impact of underlying CF-pathogenic alleles in aggregate.
Finally, the success of our de novo approach of building a phenotype risk score of CF relies on several aspects of CF. The number of CF cases in the discovery data set (71 CF cases of ~10,000 persons of European descent) has empowered our genetic association studies to reveal phenotypes that broadly cover clinical manifestations of CF. These EHR phenotypes comprise the basis for building PheRSassoc, with some being highly specific to CF (e.g., MRSA pneumonia). The availability of the CF diagnosis code in EHRs also made it easier to assign CF case status. CF is the most common recessive Mendelian disease in populations with European ancestries, and was diagnosed in ~0.1% of the patient population of our validation data set containing ~2.8 million patients. While it is unclear that CF results can be extended to rarer recessive Mendelian diseases, we believe such investigations may have value for more automated identification of patients with undiagnosed Mendelian diseases and for more complete cataloging of EHR-based phenotypic descriptions of Mendelian diseases.
The study had several limitations. First, the PheRS construction used phecodes derived from ICD billing codes. Although ICD billing codes are ubiquitous and easily shared across health systems, the mapping task from ICD codes to phecodes is not trivial and rather a growing burden. As the massive EHR data continue to accumulate, PheRS constructed using ICD codes directly would simplify the process to adopt PheRS in another health system. Second, there are correlations among the constitutive codes of PheRS that have not yet been systematically handled in the development of PheRS. Although the correlations are weak, taking into account the correlation in PheRS can further increase its performance. Third, there are individuals without cystic fibrosis who scored high (at population level) due to another disease (e.g., septicemia) when the disease manifestations (e.g., pneumonia, bacterial infection) overlap some of the scoring conditions (Supplementary Table S11–14). Future development of PheRS may consider a more sophisticated machine learning approach to find a better weighting scheme to alleviate these problems. Finally, our study suggests that PheRS could be a valuable tool to stimulate clinical suspicion of patients who may be affected by CF; however, the ultimate utility of PheRS in clinical practice would require prospective studies for further evaluation.
References
Farrell PM, White TB, Ren CL, Hempstead SE, Accurso F, Derichs N, et al. Diagnosis of cystic fibrosis: consensus guidelines from the Cystic Fibrosis Foundation. J Pediatr. 2017;181S:S4–S15 e11.
Ikpa PT, Bijvelds MJ, de Jonge HR. Cystic fibrosis: toward personalized therapies. Int J Biochem Cell Biol. 2014;52:192–200.
Rowntree RK, Harris A. The phenotypic consequences of CFTR mutations. Ann Hum Genet. 2003;67(Pt 5):471–485.
Cutting GR. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet. 2015;16:45–56.
Blackman SM, Commander CW, Watson C, Arcara KM, Strug LJ, Stonebraker JR, et al. Genetic modifiers of cystic fibrosis-related diabetes. Diabetes. 2013;62:3627–3635.
Corvol H, Blackman SM, Boelle PY, Gallins PJ, Pace RG, Stonebraker JR, et al. Genome-wide association meta-analysis identifies five modifier loci of lung disease severity in cystic fibrosis. Nat Commun. 2015;6:8382.
Wright FA, Strug LJ, Doshi VK, Commander CW, Blackman SM, Sun L, et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat Genet. 2011;43:539–546.
Chong JX, Buckingham KJ, Jhangiani SN, Boehm C, Sobreira N, Smith JD, et al. The genetic basis of Mendelian phenotypes: discoveries, challenges, and opportunities. Am J Hum Genet. 2015;97:199–215.
Castel SE, Cervera A, Mohammadi P, Aguet F, Reverter F, Wolman A, et al. Modified penetrance of coding variants by cis-regulatory variation contributes to disease risk. Nat Genet. 2018;50:1327–1334.
Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660.
Mele M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, et al. Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–665.
Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210.
Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84:362–369.
McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–1283.
Do R, Willer CJ, Schmidt EM, Sengupta S, Gao C, Peloso GM, et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat Genet. 2013;45:1345–1352.
Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529.
Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47:1091–1098.
Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31:1102–1110.
Carroll RJ, Bastarache L, Denny JC. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics. 2014;30:2375–2376.
Dodge JA, Morison S, Lewis PA, Coles EC, Geddes D, Russell G, et al. Incidence, population, and survival of cystic fibrosis in the UK, 1968-95. UK Cystic Fibrosis Survey Management Committee. Arch Dis Child. 1997;77:493–496.
Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, et al. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–1080.
Lemna WK, Feldman GL, Kerem B, Fernbach SD, Zevkovich EP, O’Brien WE, et al. Mutation analysis for heterozygote detection and the prenatal diagnosis of cystic fibrosis. N Engl J Med. 1990;322:291–296.
Stegle O, Parts L, Piipari M, Winn J, Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc. 2012;7:500–507.
Putting research data into your hands with the MarketScan Databases. 2016. http://truvenhealth.com/markets/life-sciences/products/data-tools/marketscan-databases. Accessed 2020 Feb 6.
IBM Watson Health, IBM MarketScan Research Databases 2019. https://www.ibm.com/downloads/cas/4QD5ADRL. Accessed 2020 Feb 6.
Kulaylat AS, Schaefer EW, Messaris E, Hollenbeak CS. Truven Health Analytics MarketScan Databases for clinical research in colon and rectal surgery. Clin Colon Rectal Surg. 2019;32:54–60.
Quint J. Health research data for the real world: the MarketScan database. Ann Arbor, MI: Truven Health Analytics; 2015.
Jia G, Li Y, Zhang H, Chattopadhyay I, Boeck Jensen A, Blair DR, et al. Estimating heritability and genetic correlations from large health data sets in the absence of genetic data. Nat Commun. 2019;10:5508.
Noroski L, Das S, Hajjar J. Case 40-2018: a woman with recurrent sinusitis, cough, and bronchiectasis. N Engl J Med. 2019;380:1383.
McCloskey M, Redmond AO, Hill A, Elborn JS. Clinical features associated with a delayed diagnosis of cystic fibrosis. Respiration. 2000;67:402–407.
Gan KH, Geus WP, Bakker W, Lamers CB, Heijerman HG. Genetic and clinical features of patients with cystic fibrosis diagnosed after the age of 16 years. Thorax. 1995;50:1301–1304.
Rodman DM, Polis JM, Heltshe SL, Sontag MK, Chacon C, Rodman RV, et al. Late diagnosis defines a unique population of long-term survivors of cystic fibrosis. Am J Respir Crit Care Med. 2005;171:621–626.
Bastarache L, Hughey JJ, Hebbring S, Marlo J, Zhao W, Ho WT, et al. Phenotype risk scores identify patients with unrecognized Mendelian disease patterns. Science. 2018;359:1233–1239.
Bastarache L, Bastarache JA, Denny JC. Case 40-2018: a woman with recurrent sinusitis, cough, and bronchiectasis. N Engl J Med. 2019;380:1382–1383.
Schram CA. Atypical cystic fibrosis: identification in the primary care setting. Can Fam Physician. 2012;58:1341–1345. e1699-1704
Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature. 2018;560:319–324.
Plasschaert LW, Zilionis R, Choo-Wing R, Savova V, Knehr J, Roma G, et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature. 2018;560:377–381.
Mulberg AE, Weyler RT, Altschuler SM, Hyde TM. Cystic fibrosis transmembrane conductance regulator expression in human hypothalamus. Neuroreport. 1998;9:141–144.
Guo Y, Su M, McNutt MA, Gu J. Expression and distribution of cystic fibrosis transmembrane conductance regulator in neurons of the human brain. J Histochem Cytochem. 2009;57:1113–1120.
Marcorelles P, Friocourt G, Uguen A, Lede F, Ferec C, Laquerriere A. Cystic fibrosis transmembrane conductance regulator protein (CFTR) expression in the developing human brain: comparative immunohistochemical study between patients with normal and mutated CFTR. J Histochem Cytochem. 2014;62:791–801.
Kowalczyk T, Pontious A, Englund C, Daza RA, Bedogni F, Hodge R, et al. Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb Cortex. 2009;19:2439–2450.
Acknowledgements
This work was funded by the National Institutes of Health (NIH) grants R01MH113362, U01HG009086, R35HG010718, R01HL122712, 1P50MH094267, and U01HL108634-01. A.R. also acknowledges support from the Defense Advanced Research Projects Agency (DARPA) Big Mechanism program under Army Research Office (ARO) contract W911NF1410333, the King Abdullah University of Science and Technology (KAUST), and a gift from Liz and Kent Dauten. BioVU and the Synthetic Derivative of Vanderbilt University Medical Center are supported by the National Center for Advancing Translational Science grant UL1TR000445 from NIH; the genotypes in BioVU used for the analyses described were funded by NIH grants RC2GM092618 and U01HG004603.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Disclosure
E.R.G. receives an honorarium from the journal Circulation Research of the American Heart Association, as a member of the Editorial Board. He performed consulting on pharmacogenetic analysis with the City of Hope/Beckman Research Institute. The other authors declare no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Zhong, X., Yin, Z., Jia, G. et al. Electronic health record phenotypes associated with genetically regulated expression of CFTR and application to cystic fibrosis. Genet Med 22, 1191–1200 (2020). https://doi.org/10.1038/s41436-020-0786-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-020-0786-5
Keywords
This article is cited by
-
Limited clinical utility for GWAS or polygenic risk score for postoperative acute kidney injury in non-cardiac surgery in European-ancestry patients
BMC Nephrology (2022)
-
Discerning asthma endotypes through comorbidity mapping
Nature Communications (2022)