Abstract
Purpose
Congenital contractural arachnodactyly (CCA) is an autosomal dominant connective tissue disorder manifesting joint contractures, arachnodactyly, crumpled ears, and kyphoscoliosis as main features. Due to its rarity, rather aspecific clinical presentation, and overlap with other conditions including Marfan syndrome, the diagnosis is challenging, but important for prognosis and clinical management. CCA is caused by pathogenic variants in FBN2, encoding fibrillin-2, but locus heterogeneity has been suggested. We designed a clinical scoring system and diagnostic criteria to support the diagnostic process and guide molecular genetic testing.
Methods
In this retrospective study, we assessed 167 probands referred for FBN2 analysis and classified them into a FBN2-positive (n = 44) and FBN2-negative group (n = 123) following molecular analysis. We developed a 20-point weighted clinical scoring system based on the prevalence of ten main clinical characteristics of CCA in both groups.
Results
The total score was significantly different between the groups (P < 0.001) and was indicative for classifying patients into unlikely CCA (total score <7) and likely CCA (total score ≥7) groups.
Conclusions
Our clinical score is helpful for clinical guidance for patients suspected to have CCA, and provides a quantitative tool for phenotyping in research settings.
Similar content being viewed by others
INTRODUCTION
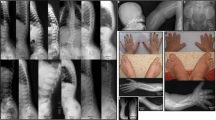
Congenital contractural arachnodactyly (CCA) or Beals syndrome (MIM 121050), is a rare autosomal dominant connective tissue disorder. The initially reported key clinical features of CCA are congenital joint contractures, arachnodactyly, crumpled ears, and kyphoscoliosis.1 Further clinical series additionally reported a tall, slender stature with dolichostenomelia, micrognathia, a highly arched palate, sternal deformities, and muscle hypoplasia.2,3,4,5
CCA closely relates to Marfan syndrome (MFS) (MIM 154700). Furthermore, some clinical overlap exists with type VI collagenopathies (Bethlem myopathy [BM; MIM 158810 and 616471] and Ullrich congenital muscular dystrophy [MIM 254090 and 616470]), as well as with Loeys–Dietz syndrome (LDS) (MIM 609192, 610168, 613795, 614816, and 615582) and distal arthrogryposes.6
In contrast to MFS, CCA patients seldom show cardiovascular involvement,1 though aortic root dilation has been observed.4,5,7,8,9 Some reports also suggest an increased occurrence of mitral valve prolapse, atrial and ventricular septal defects, and interrupted aortic arch.2,3,7,10,11,12,13 Ectopia lentis (EL) was incidentally reported in early series,14,15 but never found in patients with molecularly confirmed CCA. Other ocular features include keratoconus and myopia.3,5,14 Individuals with CCA typically do not present with intellectual disability (ID), but a delay in motor development can be present due to joint contractures.5,16 The prognosis in CCA is rather favorable. Aortic root dilatation in CCA is rare and generally mild or nonprogressive without a high risk for dissection. Joint contractures usually improve and may even resolve. However, kyphoscoliosis may be progressive and eventually require surgical treatment. Ear deformities tend to become less pronounced over time, evolving to a “tram track” appearance of the helical and antihelical cruri.5
The clinical overlap between CCA and MFS reflects similarity at the pathophysiological level. Shortly after the identification of pathogenic variants in FBN1 (encoding the extracellular matrix protein fibrillin-1) in MFS,17 FBN2 (encoding the extracellular matrix protein fibrillin-2) was mapped on chromosome 5 and found mutated in CCA patients.18,19 Most FBN2 pathogenic variants cluster in a limited region of the FBN2 gene, from exon 24 through 35. This region is homologous to the region of FBN1 that harbors most pathogenic variants underlying the neonatal MFS phenotype, hence called the neonatal region.4,5,20,21,22,23,24 However, pathogenic variants outside the neonatal region have been reported.5 Overall, the detection rate of pathogenic variants remains low in CCA (27–75%), and suggests either locus heterogeneity or clinical overdiagnosis of CCA.4,5,25
Over the last decade, next-generation sequencing (NGS) has allowed faster and less expensive genetic testing. While it was expected that NGS would facilitate counseling and management, and that it would be helpful for determining the prognosis, the identification of variants of unknown significance (VUS) and incompletely understood molecular heterogeneity mitigate the straightforward use of molecular testing in clinical practice. To address these challenges, we developed a clinical scoring system and clinical criteria to support the diagnostic process in CCA and guide molecular genetic testing. Furthermore, the classification may prove beneficial for defining patient inclusion criteria in research.
MATERIALS AND METHODS
Patients and clinical data
We performed a retrospective analysis in a large cohort of clinically diagnosed CCA probands, referred to our laboratory for molecular analysis of FBN2.
For all probands, we listed the presence or absence of the ten main clinical characteristics of CCA: crumpled ears, arachnodactyly, contractures of digits, contractures of large joints, pectus deformity, dolichostenomelia (arm span to height ratio ≥1.05), (kypho)scoliosis (scoliosis is diagnosed clinically if a vertical difference of at least 1.5 cm between the ribs of the left and right hemithorax is observed upon bending forward or if a Cobb angle of at least 20° is seen on radiographs, in the presence of a horizontal upper lining of the sacrum; increased thoracolumbar kyphosis is attributed clinically by the presence of an exaggerated sagittal curvature), muscular hypoplasia, highly arched palate, and micrognathia. We further recorded the age at time of clinical assessment to evaluate age-related changes to the clinical phenotype, and cardiovascular and ophthalmological features. Probands with EL and/or ID were excluded, as these features suggest alternative diagnoses.
The clinical data were gathered from clinical checklists completed by the referring physician at the time of referral. When available, clinical pictures were used to evaluate the presence or absence of clinical features. Where there were missing data, additional information was requested from the referring physicians.
Molecular analysis
FBN2 analysis was performed using Sanger sequencing or polymerase chain reaction (PCR)-based NGS (Miseq, Illumina, San Diego, CA, USA). Sequences were compared with the wild-type sequence NM_001999.3 (Ensembl accession number ENST00000262464.8). Nucleotides were numbered starting from the first base of the initiation codon (ATG) of the complementary DNA (cDNA) reference sequence. Amino acid residues were numbered from the first methionine residue of the reference sequence.
Based on the molecular results, probands were divided into two groups: probands with a FBN2 pathogenic variant (FBN2-positive) and probands without a FBN2 pathogenic variant (FBN2-negative). The interpretation of the pathogenicity of the identified variants was based on the integration of the results of population frequencies and in silico prediction tools using the Alamut Visual decision-support software application and the Combined Annotation Dependent Depletion (CADD) framework.26 Patients with a VUS were excluded from the study.
To identify FBN2 multiexon deletions, we used a custom-made 60K connective tissue array (Agilent SureDesign array 8 × 60K) (Agilent Technologies, Santa Clara, CA, USA), comprising probes for known genes involved in heritable connective tissue syndromes. Array comparative genomic hybridization (arrayCGH) analysis of this 60K connective tissue array was performed as previously described.27 Data analysis was executed with the in-house developed software ViVar.28
The stability of the messenger RNA (mRNA) of the mutant transcripts was confirmed using cDNA. Total RNA was extracted from fibroblasts using automated Maxwell Rapid Sample Concentrator (Promega). cDNA was prepared using iScriptRM cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA, USA), and amplified and sequenced using standard PCR and Sanger sequencing.
Clinical score and statistical analysis
To design the clinical scoring system, we compared the occurrence of each of the ten clinical features in the FBN2-positive and the FBN2-negative patient groups using the Chi-square test or Fisher’s exact test. We calculated the sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy for each clinical characteristic. Based on these results, we allocated a weighted clinical score to each of the ten clinical features, with the maximum clinical score being 20 points (when all features are present).
The total clinical score was calculated for each patient. The distribution of the total clinical scores of the FBN2-positive and FBN2-negative patient group was compared using the Mann–Whitney U test. The influence of age on the total score was estimated by calculation of the coefficient of determination by simple linear regression. We determined cut-off values of the total clinical score to differentiate between likely CCA and unlikely CCA cases, based on a receiver operating characteristic (ROC) curve. Statistical analyses were performed using SPSS Statistics 25 software.
This study was approved by the Ethics Committee of Ghent University Hospital (EC 2017/0019).
RESULTS
Cohort
We performed an extensive clinical and molecular assessment in 183 probands, referred for FBN2 analysis. Nine patients were excluded because they had EL (n = 3) or ID (n = 6), suggesting an alternative diagnosis.
Molecular analysis
In 51 of the remaining 174 probands a FBN2 variant was found, comprising 42 different FBN2 variants. Three missense variants (c.976C>T [p.Pro326Ser], c.2260G>A [p.Gly754Ser], and c.4141C>A [p.His1381Asn]) and two splice variants (c.3725-14T>G [p.?] and c.4223-16T>G [p.?]) were classified as VUS, because they had a high population frequency and/or unconvincing in silico predictions of pathogenicity. The seven patients harboring these variants were excluded from further analysis.
The variants interpreted as being (likely) pathogenic were identified in 44 individuals and included 33 missense variants, 1 splice site variant, and 3 multiexon deletions. Only one variant was located outside the neonatal region. Fibroblasts were available from two of the three patients with a multiexon deletion identified with the connective tissue array. These were used for cDNA analysis, which indicated stable expression of the mutant transcript.
An overview of the variants predicted to be pathogenic is given in Table 1 and shown in Fig. 1. The results of the in silico prediction tools and the population frequencies used to evaluate pathogenicity of all FBN2 variants are listed in Supplementary Table S1.
Determination of the clinical score
Based on molecular results, probands were subdivided into 44 FBN2-positive and 123 FBN2-negative probands. An overview of the clinical characteristics is shown in Supplementary Table S2. We calculated the sensitivity, specificity, PPV, NPV, accuracy, and two-sided P value of Chi-square test (or Fisher’s exact test) for each characteristic, and proposed a weighted clinical score based on these data (see Table 2). Crumpled ears, arachnodactyly, camptodactyly and contractures of large joints were allotted 3 points each, because these features show high sensitivity, NPV, and/or a significant Chi-square test. Dolichostenomelia and pectus deformity show the best specificity and accuracy and were given a score of 2. The remaining four clinical characteristics, kyphoscoliosis, muscle hypoplasia, highly arched palate, and micrognathia were allotted a score of 1.
Figure 2a gives an overview of the distribution of the total scores calculated for the individuals in both groups. These total clinical scores appear significantly different in FBN2-positive patients compared with the FBN2-negative group (P < 0.001).
Evaluation of the clinical scoring system and design of diagnostic criteria. (a) Bar chart giving an overview of the distribution of the total scores for FBN2-positive and FBN2-negative probands. (b) Receiver operating characteristic (ROC) curve and coordinates used for determination of a clinically relevant cut-off score to differentiate between likely congenital contractural arachnodactyly (CCA) and unlikely CCA cases. (c) Scatter plot displaying the total score as a function of age at clinical evaluation for both FBN2-positive and FBN2-negative probands.
The ROC curve used for cut point analysis is shown in Fig. 2b. Based on the sensitivity and the 1 – specificity for each score, we determined a cut-off for likely CCA at a score of 7. Hence, patients were categorized as unlikely CCA patients (if total score 0–6) and likely CCA patients (if total score 7–20).
The mean age of the patients, at the time of clinical evaluation and molecular testing, was 8.51 years (standard deviation 12.17, median 1, range 60). Simple linear regression to assess the impact of age at the time of clinical evaluation on the total score established a coefficient of determination R2 of 0.012. A scatter plot displaying the total score as a function of age at clinical evaluation is given in Fig. 2c.
Other clinical features
Information on aortic root dilation and mitral valve prolapse was available for 93 (55.7%) and 91 (54.5%) respectively of the included probands. Aortic root dilation was present in 21 patients (21/93) (22.6%), all of them classified as likely CCA patients. Four (19.0%) were FBN2-positive and 17 (81.0%) were FBN2-negative. Mitral valve prolapse and/or insufficiency was reported for 17 patients (17/91) (18.7%). Again, they were all classified within the likely CCA group, and included one (5.9%) FBN2-positive patient and 16 (94.1%) FBN2-negative patients.
Information on ophthalmological features (keratoconus) was available for 66 (39.5%) of the probands. Keratoconus was diagnosed in two patients (2/66) (3%), that were both FBN2-positive and classified as likely CCA.
Of note, one FBN2-positive likely CCA patient had a congenital diaphragmatic hernia.
DISCUSSION
We provide a clinical scoring system based on weighted clinical features to facilitate the diagnostic process, and guide the use of molecular genetic testing in patients suspected to have CCA. We anticipate that the use of a scoring system will help to quantify clinical severity and hence may turn out to be a practical tool for clinicians, patients, and researchers, and may facilitate genotype–phenotype association analysis.
Good clinical practice guidelines rely on both evidence and expert opinion, combine validity, reliability, reproducibility, and clinical applicability, and should be straightforward and easy to use in a clinical setting.29 We attributed a weighted score to different clinical characteristics, based on calculated parameters derived from the prevalence of the characteristic in the FBN2-positive and FBN2-negative group. These calculations were based on data available from medical records and did not take into account missing data. Therefore, the presence of a reporting bias cannot be excluded. Nonetheless, the attributed weights correlate well with the previously reported views about major and minor clinical characteristics for CCA,4,5 supporting the validity of the scoring system. Future prospective validation should confirm this.
The proposed criteria are practical to use, and easily applicable at clinical examination. Some characteristics may be subject to interobserver variability, but we expect this to be limited when patients are assessed by experts in connective tissue disorders.
The scoring system can be applied to different age groups, since only 1.2% of the variation in the total score in our cohort appeared to result from differences in the age at clinical evaluation. This might seem contradictory to previous literature, where it has been observed that some of the clinical features become less outspoken with age. However, residual contractures and ear malformations often remain (while not being self-reported by the patient), and other manifestations including scoliosis, dolichostenomelia, and arachnodactyly may increase with age, keeping the total score overall in balance.
To determine the cut-off value discriminating between unlikely CCA patients and likely CCA patients, we reasoned that, in view of the rarity of the disorder, it is desirable not to miss the diagnosis at the expense of the specificity of the test. Indeed, while there is no major risk for aortic complications requiring an early diagnosis of CCA, certainty about the diagnosis and prognosis can be reassuring (especially in view of the differential diagnosis with other connective tissue disorders including MFS and LDS). Using a total score of 7 as cut-off identified 95.5% of the FBN2-positive patients correctly. Only two (4.5%) of the FBN2-positive patients had a score of less than 7, and were falsely categorized within the unlikely CCA group. It should be noted that these two patients had a high number of missing data, and therefore, their true total score might have been underestimated. On the other hand, the same cut-off value faces a low specificity classifying only 17.1% of the FBN2-negative patients correctly as unlikely CCA, and will result in a substantial cost for molecular analysis. However, it should be noted that the scoring system is a continuum. An increasing score will increase specificity, with a higher a priori chance that the clinical diagnosis is correct. In this regard, a score of 11 still identifies 75% of the FBN2-positive patients, and classifies almost 60% of the patients with another underlying cause as unlikely CCA. Hence the system provides diagnostic handles both in clinical and in research settings.
The significant number of FBN2-negative patients classified as likely CCA reflects the low detection rate for FBN2 pathogenic variants in patients suspected to have CCA, either due to clinical misdiagnosis or locus heterogeneity for this disorder.5
By including all probands with a possible diagnosis of CCA, our approach reflects the situation in clinical practice. Hence, thorough molecular analysis revealed an overall detection rate of pathogenic variants of 26.3%. This is comparable with previously reported detection rates using whole FBN2 gene analyses.5,25
We classified the missense variants c.976C>T (p.Pro326Ser), c.2260G>A (p.Gly754Ser), and c.4141C>A (p.His1381Asn) as VUS, mainly because of the relatively high occurrence of these variants in population databases (over 1 in 1000 in gnomAD). However, the c.2260G>A (p.Gly754Ser) variant was previously reported as a likely causal variant, because of the classical CCA phenotype of the proband in whom it was identified.5 In the absence of segregation analysis and functional evidence, and in light of the population frequency, we assume it should be considered a class 3 variant. The splice variants c.3725-14T>G (p.?) and c.4223-16T>G (p.?) were considered VUS, based on the results of in silico prediction tools and low CADD score.
In our cohort, we detected only one (likely) pathogenic variant (c.2773G>A [p.Gly925Arg]) located outside, but in close proximity to the neonatal region. This observation confirms the essential contribution of the neonatal region to the CCA phenotype. The decision to consider this variant as pathogenic is based on in silico predictions of pathogenicity (including CADD score) and the absence of this variant from population databases. Furthermore, segregation analysis confirmed the de novo occurrence of the variant in this proband.
Our data broaden the molecular spectrum in CCA, identifying three novel multiexon deletions (leading to a skip of exon 38 to 48, 43 to 48, and 45 to 48 respectively). FBN2 gene sequencing should therefore be complemented by gene dosage analysis. For at least two of these multiexon deletions, we could show that the mutant mRNA is stably expressed. Therefore, it is likely that a shorter protein is produced and incorporated into the extracellular matrix. This further supports the hypothesis of a dominant negative effect of FBN2 pathogenic variants causing CCA, and contrasts with FBN1 pathogenic variants in MFS where both dominant negative and haploinsufficiency mechanisms are implicated in the pathogenesis.5,22,30,31,32,33,34,35,36
A diagnostic flowchart designed to guide clinicians in the diagnostic decision-making process is given in Fig. 3. For patients with a score of 0–6, CCA is unlikely, thus another diagnosis should be considered and FBN2 analysis is not indicated. For patients with a score of 7–20, and thus categorized as likely CCA, molecular genetic testing of FBN2 is indicated to confirm the diagnosis. If comprehensive genetic analysis is normal, disorders resembling CCA, such as MFS and LDS, should be excluded. Where the diagnosis remains molecularly unexplained, the patients can be included in research studies to evaluate CCA locus heterogeneity. Within our cohort, seven FBN2-negative likely CCA patients were eventually diagnosed with MFS (total scores were 9, 12, 13, 13, 14, 15, and 17), one was molecularly confirmed to have LDS (total score 7), and one was confirmed to have BM (total score 8).
In patients presenting with EL and/or ID, CCA is unlikely. In case of thoracic aortic aneurysm (TAA), CCA is not excluded per se, but diagnostics should focus first on established TAA genes.37
Despite the lack of good follow-up data and the absence of unequivocal evidence of an aortic risk in CCA, the observation of cardiovascular features in likely CCA patients indicates that cardiac screening is advisable (e.g., every 3–5 years). Similarly, ophthalmological follow-up is advised in likely CCA patients.
In conclusion, we have developed a clinical scoring system and clinical diagnostic criteria for CCA, based on the presence or absence of the ten main clinical features of this disorder. This tool may prove helpful in diagnostic decision making, as well as in future CCA research.
References
Hecht F, Beals RK. “New” syndrome of congenital contractural arachnodactyly originally described by Marfan in 1896. Pediatrics. 1972;49:574–579.
Ramos Arroyo MA, Weaver DD, Beals RK. Congenital contractural arachnodactyly. Report of four additional families and review of literature. Clin Genet. 1985;27:570–581.
Viljoen D. Congenital contractural arachnodactyly (Beals syndrome). J Med Genet. 1994;31:640–643.
Gupta PA, Putnam EA, Carmical SG, et al. Ten novel FBN2 mutations in congenital contractural arachnodactyly: delineation of the molecular pathogenesis and clinical phenotype. Hum Mutat. 2002;19:39–48.
Callewaert BL, Loeys BL, Ficcadenti A, et al. Comprehensive clinical and molecular assessment of 32 probands with congenital contractural arachnodactyly: report of 14 novel mutations and review of the literature. Hum Mutat. 2009;30:334–341.
Bamshad M, Jorde LB, Carey JC. A revised and extended classification of the distal arthrogryposes. Am J Med Genet. 1996;65:277–281.
Macnab AJ, D’Orsogna L, Cole DE, et al. Cardiac anomalies complicating congenital contractural arachnodactyly. Arch Dis Child. 1991;66:1143–1146.
Gupta PA, Wallis DD, Chin TO, et al. FBN2 mutation associated with manifestations of Marfan syndrome and congenital contractural arachnodactyly. J Med Genet. 2004;41:e56.
Takeda N, Morita H, Fujita D, et al. Congenital contractural arachnodactyly complicated with aortic dilatation and dissection: case report and review of literature. Am J Med Genet A. 2015;167a:2382–2387.
Anderson RA, Koch S, Camerini-Otero RD. Cardiovascular findings in congenital contractural arachnodactyly: report of an affected kindred. Am J Med Genet. 1984;18:265–271.
Bell RE, Wheller JJ. Cardiac defects in a patient with congenital contractural arachnodactyly. South Med J. 1985;78:742–743.
Huggon IC, Burke JP, Talbot JF. Contractural arachnodactyly with mitral regurgitation and iridodonesis. Arch Dis Child. 1990;65:317–319.
Su PH, Hou JW, Hwu WL, et al. Congenital contractural arachnodactyly (Beals syndrome). Acta Paediatr Taiwan. 2000;41:59–62.
Bass HN, Sparkes RS, Crandall BF, Marcy SM. Congenital contractural arachnodactyly, keratoconus, and probable Marfan syndrome in the same pedigree. J Pediatr. 1981;98:591–593.
Bawle E, Quigg MH. Ectopia lentis and aortic root dilatation in congenital contractural arachnodactyly. Am J Med Genet. 1992;42:19–21.
Tuncbilek E, Alanay Y. Congenital contractural arachnodactyly (Beals syndrome). Orphanet J Rare Dis. 2006;1:20.
Dietz HC, Cutting GR, Pyeritz RE, et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337–339.
Lee B, Godfrey M, Vitale E, et al. Linkage of Marfan syndrome and a phenotypically related disorder to two different fibrillin genes. Nature. 1991;352:330–334.
Putnam EA, Zhang H, Ramirez F, Milewicz DM. Fibrillin-2 (FBN2) mutations result in the Marfan-like disorder, congenital contractural arachnodactyly. Nat Genet. 1995;11:456–458.
Putnam EA, Cho M, Zinn AB, et al. Delineation of the Marfan phenotype associated with mutations in exons 23-32 of the FBN1 gene. Am J Med Genet. 1996;62:233–242.
Park ES, Putnam EA, Chitayat D, et al. Clustering of FBN2 mutations in patients with congenital contractural arachnodactyly indicates an important role of the domains encoded by exons 24 through 34 during human development. Am J Med Genet. 1998;78:350–355.
Robinson PN, Godfrey M. The molecular genetics of Marfan syndrome and related microfibrillopathies. J Med Genet. 2000;37:9–25.
Faivre L, Collod-Beroud G, Loeys BL, et al. Effect of mutation type and location on clinical outcome in 1,013 probands with Marfan syndrome or related phenotypes and FBN1 mutations: an international study. Am J Hum Genet. 2007;81:454–466.
Faivre L, Collod-Beroud G, Callewaert B, et al. Clinical and mutation-type analysis from an international series of 198 probands with a pathogenic FBN1 exons 24-32 mutation. Eur J Hum Genet. 2009;17:491–501.
Nishimura A, Sakai H, Ikegawa S, et al. FBN2, FBN1, TGFBR1, and TGFBR2 analyses in congenital contractural arachnodactyly. Am J Med Genet A. 2007;143a:694–698.
Kircher M, Witten DM, Jain P, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315.
Menten B, Buysse K, Vandesompele J, et al. Identification of an unbalanced X-autosome translocation by array CGH in a boy with a syndromic form of chondrodysplasia punctata brachytelephalangic type. Eur J Med Genet. 2005;48:301–309.
Sante T, Vergult S, Volders PJ, et al. ViVar: a comprehensive platform for the analysis and visualization of structural genomic variation. PLoS ONE. 2014;9:e113800.
Steinbrook R. Guidance for guidelines. N Engl J Med. 2007;356:331–333.
Milewicz DM, Pyeritz RE, Crawford ES, Byers PH. Marfan syndrome: defective synthesis, secretion, and extracellular matrix formation of fibrillin by cultured dermal fibroblasts. J Clin Invest. 1992;89:79–86.
Dietz HC, McIntosh I, Sakai LY, et al. Four novel FBN1 mutations: significance for mutant transcript level and EGF-like domain calcium binding in the pathogenesis of Marfan syndrome. Genomics. 1993;17:468–475.
Eldadah ZA, Brenn T, Furthmayr H, Dietz HC. Expression of a mutant human fibrillin allele upon a normal human or murine genetic background recapitulates a Marfan cellular phenotype. J Clin Invest. 1995;95:874–880.
Dietz HC, Pyeritz RE. Mutations in the human gene for fibrillin-1 (FBN1) in the Marfan syndrome and related disorders. Hum Mol Genet. 1995;4:1799–1809.
Milewicz DM, Urban Z, Boyd C. Genetic disorders of the elastic fiber system. Matrix Biol. 2000;19:471–480.
Judge DP, Biery NJ, Keene DR, et al. Evidence for a critical contribution of haploinsufficiency in the complex pathogenesis of Marfan syndrome. J Clin Invest. 2004;114:172–181.
Dietz HC, Loeys B, Carta L, Ramirez F. Recent progress towards a molecular understanding of Marfan syndrome. Am J Med Genet C Semin Med Genet. 2005;139c:4–9.
Renard M, Francis C, Ghosh R, et al. Clinical validity of genes for heritable thoracic aortic aneurysm and dissection. J Am Coll Cardiol. 2018;72:605–615.
Acknowledgements
B.C. is a senior clinical investigator funded by Fund for Scientific Research Flanders (FWO). This project was funded by the Special Research foundation of Ghent University to B.C. (starting grant BOFSTA2016002401) and to A.D.P. (Methusalem grant BOF15/MET-V/011). We thank Jan C. Oosterwijk and Rebecca Igbokwe for their helpful comments on the manuscript. We are grateful to all patients and their families for their interest and participation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
The authors declare no conflicts of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Meerschaut, I., De Coninck, S., Steyaert, W. et al. A clinical scoring system for congenital contractural arachnodactyly. Genet Med 22, 124–131 (2020). https://doi.org/10.1038/s41436-019-0609-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-019-0609-8