Abstract
Purpose
ABCA4-associated disease, a recessive retinal dystrophy, is hallmarked by a large proportion of patients with only one pathogenic ABCA4 variant, suggestive for missing heritability.
Methods
By locus-specific analysis of ABCA4, combined with extensive functional studies, we aimed to unravel the missing alleles in a cohort of 67 patients (p), with one (p = 64) or no (p = 3) identified coding pathogenic variants of ABCA4.
Results
We identified eight pathogenic (deep-)intronic ABCA4 splice variants, of which five are novel and six structural variants, four of which are novel, including two duplications. Together, these variants account for the missing alleles in 40.3% of patients. Furthermore, two novel variants with a putative cis-regulatory effect were identified. The common hypomorphic variant c.5603A>T p.(Asn1868Ile) was found as a candidate second allele in 43.3% of patients. Overall, we have elucidated the missing heritability in 83.6% of our cohort. In addition, we successfully rescued three deep-intronic variants using antisense oligonucleotide (AON)-mediated treatment in HEK 293-T cells and in patient-derived fibroblast cells.
Conclusion
Noncoding pathogenic variants, novel structural variants, and a common hypomorphic allele of the ABCA4 gene explain the majority of unsolved cases with ABCA4-associated disease, rendering this retinopathy a model for missing heritability in autosomal recessive disorders.
Similar content being viewed by others
INTRODUCTION
Stargardt disease (STGD1, MIM 248200) is one of the most frequent inherited retinal diseases (IRDs), with an estimated prevalence of 1/8000–1/10,000. The underlying disease gene for STGD1 is the large ATP-binding cassette subfamily A member 4 gene (ABCA4, MIM 601691), consisting of 50 coding exons.1 Apart from STGD1, a juvenile macular dystrophy, biallelic ABCA4 variants have been linked to a spectrum of autosomal recessive IRD phenotypes, varying from cone-rod dystrophy (CRD), atypical retinitis pigmentosa (RP), fundus flavimaculatus, generalized choriocapillaris dystrophy (GCCD), to rapid-onset chorioretinopathy (ROC), jointly named ABCA4-associated disease.2,3,4,5 A genotype–phenotype correlation model was proposed to explain this large variety of phenotypes attributed to biallelic pathogenic variants in ABCA4, linking the amount of residual activity of ABCA4 to the corresponding phenotype. According to this model, patients with two severe variants (null alleles) present with atypical RP or ROC while a severe variant in trans to a moderate variant gives rise to CRD, and a severe and mild variant or two moderate variants give rise to STGD1 (refs. 6,7). The variant spectrum of ABCA4-associated disease is characterized by vast allelic heterogeneity, with over 800 variants identified (https://databases.lovd.nl/shared/genes/ABCA4), the majority of which are located in the coding region of ABCA4. Furthermore, a large number of intronic noncanonical splice site variants have been identified, with c.5461–10T>C being the third most frequent pathogenic variant.8,9 Previous studies indicate that copy-number variations (CNVs) represent only a minor fraction of the variant spectrum.3,7,10,11,12,13,14,15,16,17,18 This large number of (likely) pathogenic variants in ABCA4 can only explain 60–75% of STGD1 and other monoallelic ABCA4-associated disease cases, suggesting missing heritability in noncoding regions. Indeed, we and others uncovered noncoding deep-intronic variants as second pathogenic alleles.10,19,20,21,22 Braun et al.19 and Zernant et al.20 identified a subset of noncoding variants with a presumed effect on splicing. We found the deep-intronic variant c.4539+2001G>A (denoted as V4) as the second missing allele in more than 25% of Belgian monoallelic STGD1 patients, representing the first noncoding founder pathogenic variant in ABCA4 (ref.21). Schulz et al.22 analyzed the coding region of ABCA4 and parts of intron 30 and 36, known to harbor deep-intronic variants, in a large (p = 335) German STGD1 cohort. They identified rare deep-intronic variants in only four patients and reported six putative risk-modulating common variants, one of which is the hypomorphic recurrent ABCA4 variant c.5603A>T p.(Asn1868Ile) (minor allele frequency [MAF] 7%, Genome of the Netherlands database [GoNL]). Zernant et al.23 demonstrated that this variant contributes to 10% of the ABCA4 disease alleles, representing the second missing allele in over 50% of monoallelic STGD1 cases, often displaying a late-onset phenotype with foveal sparing.
Noncoding deep-intronic splice variants represent potential therapeutic targets for approaches such as antisense oligonucleotide (AON)-mediated rescue or CRISPR/Cas9-mediated correction of deep-intronic pathogenic variants, as illustrated for CEP290, USH2A, OPA1, and recently for ABCA4, emphasizing the relevance of this study.24,25,26,27
Taken together, this prompted us to elucidate the missing heritability in a cohort of 67 Belgian and German patients diagnosed with ABCA4-associated disease, with only one heterozygous (p = 64) or no (p = 3) identified ABCA4 pathogenic variant after first-line screening.
MATERIALS AND METHODS
Patient cohort
Patients consented to this study, which adhered to the tenets of the Declaration of Helsinki (protocols B670201525349 and B670201734438). All individuals had a clinical diagnosis of an ABCA4-associated disease, such as STGD1 (Table S1), and underwent testing of the coding region of ABCA4, revealing either one pathogenic variant (p = 64) or no pathogenic variant (p = 3) (Figure S1). Info on clinical assessments and genotype–phenotype correlations can be found in Supplemental Methods.
Locus enrichment
Using a custom HaloPlex Target Enrichment kit (Agilent, Santa Clara, CA, USA), a conserved block of synteny encompassing the ABCA4 gene (chr1:94337885-94703604, GrCh37/hg19 assembly) was enriched.28 Approximately 98% of the region could be covered, while gaps often corresponded with repeat-rich areas (Figure S2). The locus and the entire ABCA4 gene were sequenced with an average coverage depth of 263.6× and 343.4×, respectively.
Next-generation sequencing and data analysis
Samples underwent targeted next-generation sequencing (NGS) (MiSeq, Illumina, CA, USA). Read mapping, single-nucleotide variant, structural variant, and indel calling and subsequent variant identification and annotation were performed using CLC Bio Software (CLC Bio Genomics Workbench; QIAGEN, Germany). Annotations included frequencies from a subset of the UK National Institute for Health Research (NIHR) BioResource Rare Diseases Whole Genome Sequencing data set containing 7322 individuals without a known IRD, the Genome Aggregation Database (gnomAD) and GoNL databases, presence within published minor exons of ABCA4 or 200 bp of neighbouring sequence (Table S2), overlap with long noncoding RNAs (lncRNAs) (Table S2) and published noncoding ABCA4 variants (Table S3), and presence in candidate regulatory regions, as determined by ATAC-seq, RNA-seq, H3K27Ac, and H3K4me2 ChIP-seq data generated on adult human retina (Table S4; unpublished data: Cherry et al.,doi.org/10.1101/412361).19,20,21,22,29 Further annotation was performed via Alamut Batch (Interactive Biosoftware, France). A custom Lua script was used for additional data processing.
ABCA4 variant analysis
First, variants within ABCA4 (NM_00350.2) were investigated. After identifying known pathogenic variants, only variants with MAF ≤1% in the aforementioned population databases were selected. Per individual, all remaining rare noncoding variants within ABCA4 were analyzed for predicted effects on splice sites, exonic splice enhancers (ESEs), and splice silencers using splice annotations of Alamut Batch, Alamut Visual (Interactive Biosoftware, France), and Human Splicing Finder (http://www.umd.be/HSF3/). Second, variants located within putative regulatory regions in the enriched locus were selected and annotated (unpublished data, Cherry et al., https://doi.org/10.1101/412361, RegulomeDB). In the absence of candidate variants, rare ABCA4 variants outside of the regulatory regions were also investigated. Polymerase chain reaction (PCR) primers for validation are listed in Table S5. If possible, segregation results were used for filtering.
In vitro splicing assays
Minigenes
Genomic DNA from patients who are heterozygous carriers of putative splice variants was used for creating wild-type (WT) and mutant minigenes for seven variants (Table S6); minigene assays were performed as described.30 Primers and conditions are listed in Table S7.
Midigenes
We used the midigene library and protocol previously described to create WT and mutant midigenes for six variants and to perform midigene assays (Tables S6 and S8) (ref. 30). Transfection was performed in triplicate in HEK 293-T cells.
AON-mediated rescue experiments
AON design
Per variant, AONs with a phosphorothioate backbone with a 2-O-methyl sugar modification (2OME/PS) were designed, as described (Table S9) (ref. 31). In addition, a sense oligonucleotide (SON) was synthesized. Synthesis was done by Eurogentec (Belgium). An AON concentration of 0.5 µM was used.
In vitro rescue studies in transfected HEK 293-T cells
HEK 293-T cells were transfected with 1.5 µg of WT or mutant midigene construct. After 24 hours, each well was trypsinized and subdivided in 5 wells of a 24-well plate. After reattachment, cells were transfected with the AON, the SON, or not treated (NT) (FuGENE HD Transfection Reagent, Promega, The Netherlands). After 48 hours, RNA was isolated (NucleoSpin RNA kit, Macherey-Nagel, Germany) followed by complementary DNA (cDNA) synthesis (iSCRIPT cDNA synthesis kit, Bio-Rad, Belgium). A PCR with 50 ng of cDNA was performed of which 10 µl was loaded on a 2% agarose gel (primers: Table S10). Rhodopsin was used as transfection and loading control. Two independent experiments were performed. Quantification of transcript ratios was done via a Fragment Analyzer Auto Capillary Electrophoresis System (Advanced Analytical Technologies, Inc., France).
In vitro rescue studies in patient-derived fibroblast cells
Patient’s fibroblasts (c.4539+1106C>T) were seeded and transfected with either 0.5 µM of AON, SON, or empty liposomes (NT). Cycloheximide (CHX) was added as a nonsense-mediated decay (NMD) blocker. Four hours later RNA was isolated followed by cDNA synthesis (Invitrogen SuperScript IV VILO Master Mix, Thermo Fisher Scientific, Belgium). For each PCR, 80 ng of cDNA was used, except for actin (50 ng) (primers: Table S10). Then, 20 µl of each ABCA4 reaction was loaded onto a 2% agarose gel using 10 µl of the actin reaction as loading control. Experiments were performed in two independent replicates. Quantification of the transcripts was performed using the capillary electrophoresis system described above.
In vitro dual-luciferase assays
Constructs and assays
Inserts were PCR amplified using genomic reference DNA (Roche) and primers containing restriction sites (Table S11). After restriction, the insert was cloned into a pgl4.23 or pgl4.10 vector (Promega, The Netherlands) and mutagenized (Q5-Site-Directed Mutagenesis Kit, New England Biolabs, France). hTert RPE-1 cells (ATCC, VA, USA) were transfected with 1.5 µg WT or mutant luciferase construct and a Renilla reporter vector (pRL, Promega, The Netherlands) (Lipofectamine 3000, Thermo Fisher Scientific, Belgium). Experiments were performed in triplicate, and a minimum of six independent experiments were performed for five variants (Table S12 and Figure S3). Assays were performed using the Dual-Luciferase Reporter Assay system and Glomax 96 microplate luminometer (Promega, The Netherlands).
Statistical data analysis
A hierarchical generalized linear mixed model (HGLMM) was fitted to the normalized relative luminometer unit values (normalized luciferase/Renilla RLU ratios). Additional information can be found in Supplemental Methods.
CNV analysis
Locus resequencing NGS data of all patients was mapped with Burrows–Wheeler Aligner (BWA) against the hg19 human reference genome, followed by local realignment of reads with Genome Analysis Toolkit (GATK) and CNV detection using the R package ExomeDepth.32,33,34 CNVs were annotated with ANNOVAR.35 Furthermore, an in-house developed customized array comparative genomic hybridization (CGH) platform (arrEYE) was used for high-resolution CNV analysis of coding and noncoding regions of ABCA4 and other retinal disease genes in a subset of ten patients (Figure S4) (ref. 36). Data was analyzed using the ViVar platform (https://vivar.cmgg.be). CNVs were confirmed and delineated using (junction) PCRs (Supplemental Methods).
Assessment of the underlying mechanisms of the identified CNVs
For each CNV an extensive bioinformatics analysis was performed as previously described37 (Supplemental Methods). Microhomology at the breakpoints was assessed using ClustalW (Figure S5). If both breakpoints of a CNV overlap with a repetitive element, the consensus sequence was retrieved from the Dfam database and sequence identity between the repetitive elements was determined using BLAST2. The potential for formation of non-B DNA conformations in the breakpoint regions was examined using the non-B DNA motif search tool (nBMST); and QGRS Mapper. Fuzznucc was used to investigate the presence of 40 sequence motifs (Tables S13 and S14).
RESULTS
Enrichment and targeted resequencing of the ABCA4 locus
Targeted resequencing revealed 5840 variants with MAF ≤1% in 67 patients (87 variants/patient) located within the sequenced region (365 kb) of which 439 variants are located within ABCA4 (6.5 variants/patient). Elimination of known pathogenic variants and certain variants in cis with these variants further reduced the number of candidate pathogenic variants to 121 variants, occurring 169 times (2.5 variants/patient) (Table S15). Thirteen candidate splice variants were selected for analysis via splice assays (Table S6).
Identification and functional assessment of novel noncoding (deep-)intronic splice variants
Summary
Overall, a splice effect was demonstrated by in vitro splice assays for nine intronic variants (n = 24) found in 21 monoallelic patients and 2 patients without prior identified pathogenic variants: c.161–23T >G (n = 1), c.769–784C >T (n = 1), c.859–540C>G (n = 1), c.3191–11T>A (n = 1), c.4253+43G>A (n = 8), c.4539+1106C>T (n = 2), c.4539+2001G>A (n = 7), c.4539+2064C>T (n = 2), and c.5197–557G>T (n = 1) (Fig. 1, Fig. 2, Table 1, and Table S1). Splice predictions are listed in Table S16. Splice assays for c.769–784C>T, c.4253+43G>A, and c.4539+2001G>A, also found in a Dutch cohort using the same resequencing approach, are reported by Sangermano et al. (this issue) and Albert et al. (ref. 26,38).
Minigene and midigene results for intronic ABCA4 splice variants. For each depicted variant (a-f) an agarose gel image is shown on the left, representing the reverse transcription polymerase chain reaction (RT-PCR) product derived from the mutant (MT) and the respective wild-type (WT) construct. On the right, the corresponding cartoon of observed sequences is shown, with mutant at the top and wild type below (exception for variant a). Above the sequence, a cartoon of the minigene or midigene construct is presented; arrows depict the location of the primers used for RT-PCR. A black “x” represents the location of the variant. “E” stands for exon, flanking Rhodopsin (RHO) exons are depicted in white, ABCA4 exons in gray, and insertions in red. “E30 M” denotes the full length exon 30, as described in NM_00350.2 (hg19), “E30 S” refers to a smaller exon 30, lacking the last 73 nucleotides p.(Cys1490Glufs*12), which was observed in several transcripts.
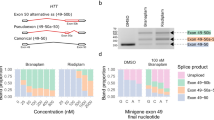
Antisense oligonucleotide (AON)-mediated rescue for intronic ABCA4 splice variants. For three deep-intronic ABCA4 variants (c.859–540C>G, c.5197–557G>T, and c.4539+1106C>T), AON rescue experiments on transfected HEK 293-T cells were undertaken (a–c). AON experiments were also performed on the fibroblasts of a patient with c.[4539+1106C>T];[6089G>A] and control fibroblasts (d). A gel image is shown on the left, depicting the reverse transcription polymerase chain reaction (RT-PCR) product derived from mutant (MT) and wild-type (WT) midigene construct after transfection with three different AONs (HEK: untransfected HEK 293-T, MQ: negative control PCR, NT: nontransfected cells, SON: sense oligonucleotide). Rhodopsin (RHO) and actin (ACTB) were used as loading controls for RT-PCR products derived from experiments on HEK 293-T cells and fibroblasts, respectively. In fibroblasts, cycloheximide (CHX) was added. On the right, resulting RT-PCR products after AON rescue experiments are semiquantified using capillary analysis, using the ratio WT transcript/transcript including pseudo-exon (PE). If multiple PE containing transcripts were formed, they are grouped together in one PE group.
Three variants near coding exons of ABCA4
Three splice variants are located within 50 bp of a canonical splice site of ABCA4. Both c.161–23T>G (intron 2) and c.4253+43G>A (intron 28) barely reduce the strength of the nearest canonical splice site but have a predicted effect on ESE binding and break silencer motifs. Splice assays reveal partial exon skipping for both variants (Fig. 1 and Sangermano et al. this issue), predicted to induce a frameshift (Table 1). [38] The c.161–23T>G variant occurs in a patient who carries the complex allele c.[2588G>C;5603A>T] (p.[Gly863Ala, Gly863del; Asn1868Ile]) in trans (Table S1) (ref. 8). In 5/8 patients c.4253+43G>A is likely in cis with c.6006–609T>A, a variant for which no splice effect could be observed (Sangermano et al. this issue).20,38 Segregation of this complex allele in trans with the pathogenic allele c.5461–10T>C in affected siblings (p = 2) as well as in an unaffected parent in the same family, and in a homozygous state in an unaffected sibling supports a hypomorphic nature of this complex allele (Figure S6). The c.3191–11T>A variant weakens the canonical acceptor splice site (ASS) of exon 22 and creates a strong novel ASS, leading to a 9-bp insertion in the mutant transcript as observed by minigene assays. This variant co-occurs with c.6089G>A p.(Arg2030Gln) in a STGD1 patient.8
Four deep-intronic variants strengthen a cryptic splice site
The deep-intronic variants c.859–540C>G (intron 7), c.4539+1106C>T (intron 30), and c.5197–557G>T (intron 36) create or strengthen cryptic donor splice sites (DSSs) while c.769–784C>T (intron 6) impacts an ASS.
The DSSs created or strengthened by c.859–540C>G, c.4539+1106C>T, and c.5197–557G>T are located downstream to a strong cryptic ASS, generating pseudo-exons (PEs) of 141 bp, 68 bp, and 188 bp respectively, as confirmed via midigene assays (Fig. 1, Table 1). Furthermore, c.4539+1106C>T creates another, less abundant PE (112 bp) due to the usage of another cryptic ASS (Fig. 1, Table 1). All included PEs harbor a premature termination codon (PTC), most likely resulting in the aberrant transcript undergoing NMD.
Specifically, c.859–540C>G occurs in trans with c.4539+2001G>A (V4) in a patient without prior identified ABCA4 pathogenic variants. The c.4539+1106C>T variant was identified in two unrelated STGD1 patients, in trans with either c.6089G>A; p.(Arg2030Gln) or c.5882G>A; p.(Gly1961Glu).
The c.5197–557G>T variant is located in known minor exon 36.2, in which no variants have been yet identified, as opposed to minor exon 36.1 (Figure S7) (ref. 19). Here it was found in trans with c.5917del p.(Val1973*).
Variant c.769–784C>T (intron 6) strengthens an ASS, upstream to a strong cryptic DSS, inducing a PE (162 bp) in only a minority of messenger RNA (mRNA) from the mutant construct (Table 1, Sangermano et al. this issue).38 Because this variant likely occurs in cis with the hypomorphic c.5603A>T p.(Asn1868Ile) in the genotype c.[1454del];[769–784C>T;5603A>T], the pathogenicity of this variant is currently considered unclear.
Two deep-intronic variants affect splice enhancers and/or silencers
Variant c.4539+2001G>A (V4) (n = 7) is a known Belgian founder variant leading to a recently described PE inclusion (345 bp), similar to c.4539+2028C>T (V5) (refs. 21,26). Variant c.4539+2064C>T (n = 2) is located within the same PE. Both have a predicted effect on ESE binding but while V4 is predicted to create ESEs and abolish splice silencers within this PE, c.4539+2064C>T is predicted to create a novel silencer within this PE. Splice assays for the latter revealed several aberrant transcripts, two including the reported PE.26
AON-mediated rescue in HEK 293-T cells and patient fibroblasts
AON-mediated rescue was undertaken for c.859–540C>G, c.4539+1106C>T, and c.5197–557G>T. Three different AONs were designed per variant, aimed at blocking ESE motifs and excluding the PE (Fig. 2, Table S9). Rescue experiments were conducted in transfected HEK 293-T cells for all three variants, and in patient-derived fibroblasts for c.4539+1106C>T.
The effect of c.859–540C>G could be fully abolished by two AONs, and partially by AON3 (43.25%) (Fig. 2). For c.5197–557G>T, all three AONs were able to restore normal splicing, with no detection of the mutant band in the AON1- and AON2-rescue (Fig. 2). AON-mediated rescue of c.4539+1106C>T was undertaken in transfected HEK 293-T cells and patient-derived fibroblast cells (c.[4539+1106C>T];[6089G>A]). In HEK 293-T cells AON1 and AON2 restored the amount of normal transcript, while AON3 retained PE inclusions in the majority (86.7%) of the transcripts. In fibroblasts, AON2 showed the highest efficacy. AON-mediated correction of c.769–784C>T and c.4253+43G>A, can be found in Sangermano et al. (this issue), while AON rescue of the aberrant transcript caused by c.4539+2001G>A (V4) was recently reported.26,38
Filtering and functional analysis of putative cis-regulatory variants
After functional analysis of 13 candidate splice variants, remaining variants were assessed for their presence within or near candidate regulatory regions within or flanking ABCA4 (Table S4; unpublished data:, Cherry et al., doi.10.1101/412361). After filtering, eight variants located within candidate regulatory regions remained, four of which were investigated using dual-luciferase reporter assays. Two additional variants outside of these regions were also selected because they were the only remaining rare candidate variants and are within predicted transcription factor binding sites (TFBSs). An overview of these six variants can be found in Table S12.
After reporter assays, two variants showed significant downregulation of luciferase expression of mutant to WT transfected RPE-1 cells: c.2919–383C>T (n = 1) and c.768+3223C>T (n = 1), with an average reduction of the normalized (luciferase/Renilla) relative light unit (RLU) values of 36.9% (p = 0.004) and 32.3% (p < 0.001) respectively (Figure S3). Variant c.2919–383C>T is located within a predicted ZNF143/STAF binding site, 18 bp adjacent to a predicted regulatory region (OREG0013611, chr1:94510701-94511661) with CTCF binding. It occurs in the following genotype c.[5461–10T>C;5603A>T];[2919–383C>T;5603A>T], sufficient to cause the late-onset phenotype observed and thus categorized as a potential modifying variant. Variant c.768+3223C>T is located within a regulatory region that shows moderate WT activity within adult human retina (unpublished data, Cherry et al., doi.org/10.1101/412361). The variant is predicted to overlap with TFBSs such as RBL2 (PAZAR data set) and has predicted binding sites for FOXP2, POLR2A, and REST proteins (ChIP-Seq data, RegulomeDB). As the variant occurs in a patient without a coding ABCA4 pathogenic variant, the implication of another disease gene cannot be ruled out (Figure S6).
Identification and characterization of structural variants implicating ABCA4
NGS-based CNV analysis was performed for all 67 patients. Furthermore, customized array-based high-resolution CNV analysis was used to investigate ten patients without a clear pathogenic noncoding sequence variant.36 We identified six CNVs involving ABCA4, four of which are novel (Fig. 3, S4, and S8; Table 2). A 5305-bp deletion of exon 5 was found in one monoallelic patient, c.442+799_c.570+541del. Exon 5 deletions have been previously reported in a Dutch STGD1 cohort.3,10 A novel deletion spanning exons 10–11 was identified in one patient, c.1239+291_1555-5574del (10,140 bp). This predicted in-frame deletion occurs in trans with c.122G>A; p.(Trp41*). A 4024-bp deletion of exons 20–22 was identified (c.2918+775_3328+640del), revealing a known recurrent deletion.3,7,10 Another novel deletion (exons 40–50) was identified and characterized at the nucleotide level, c.5585–166_*1254del (19,112 bp). It eliminates the 3′UTR of the mRNA, most likely rendering the resulting transcript unstable and occurs in trans with the complex allele c.[2588G>C;5603A>T] (p.[Gly863Ala, Gly863del;Asn1868Ile]).
Schematic overview of the ABCA4 deletions and duplications identified in this study. ABCA4 exons involved in the deletions (a-d) and duplications (e, f) identified in this study are visualized in red and blue, respectively. The localization of the primers used to perform a junction polymerase chain reaction (PCR) and to delineate the copy-number variants (CNVs) are depicted as black arrows; nucleotides on the cartoons represent microhomology at the boundaries. The orientation of the tandem duplications toward ABCA4 is the most likely one based on (non)amplification of several junction regions (data not shown). InsA: insertion of an A at a CNV junction.
A duplication of over 26 kb (exons 2–6) was identified, starting within the first intron of ABCA4, c.67–975_769-4582dup{insA}. At the junction an information scar (insertion of an A) could be found. This in-frame tandem duplication is found in trans with the missense variant c.5381C>A; p.(Ala1794Asp) in a patient. Another duplication located within intron 1 was identified in a STGD1 patient carrying the c.5714+5G>A allele (Table S1). Characterization of the junction showed a tandem noncoding duplicated region of 7006 bp, c.66+520_67-389dup.
A bioinformatics analysis reveals microhomology at the breakpoints of all deletions and the non-coding duplication, corresponding with a replicative-based CNV mechanism (Figure S5). Duplication c.67-975_769-4582dup{insA} is likely due to nonhomologous end joining (NHEJ), as there is both microhomology and an information scar at the junction (Tables S13 and S14).
The hypomorphic ABCA4 allele c.5603A>T as a second pathogenic variant
After removing patients solved with pathogenic splice or structural variants (27/67, 40.3%), 40 patients remained unsolved (Figure S1). The hypomorphic c.5603A>T p.(Asn1868Ile) variant was recently found to be the potential second missing allele in over 50% of monoallelic STGD1 cases.23 When in trans to a complete loss-of-function ABCA4 allele, this variant was shown to often lead to a late-onset phenotype. Here, it occurs as a possible trans allele in 29 of the remaining 40 patients (72.5%), even in a few cases that do not strictly meet all three criteria proposed: (1) late age of onset (>35 years), (2) complete loss-of-function of the first allele, and (3) foveal sparing. The segregation of this hypomorphic variant in trans with a pathogenic variant is clear in 50% of the cases; for the remaining patients this is undetermined (Table S1). While the average age of onset (AOO) of the patients solved with pathogenic deep-intronic splice variants and CNVs is 22.5 years, the average AOO of these 29 patients is 41 years, with 19/29 patients having an AOO >35 years. These findings suggest that c.5603A>T can be considered to be the missing allele in up to 43.3% (29/67) of the initial cohort.
DISCUSSION
In this study, we aimed to elucidate the missing heritability in a cohort of molecularly unsolved STGD1 patients using a locus-specific analysis targeting a syntenic region comprising ABCA4 and its putative cis-regulatory domain.28 This approach led to a molecular diagnosis in 83.6% (56/67) of patients. Noncoding splice variants and distinct CNVs account for the missing allele in 31.3% (21/67) and 9% (6/67) respectively, while the hypomorphic variant c.5603A>T likely contributes to another 43.3% (29/67) of the missing alleles. Two cis-regulatory variants and one deep-intronic splice variant with potential modifying effect were identified in three patients.
In the category of noncoding splice variants, nine distinct variants were found, six of which were novel (c.161–23T>G, c.769–784C>T, c.859–540C>G, c.3191–11T>A, c.4539+1106C>T, and c.5197–557G>T) and three previously described as (possibly) disease-associated (c.4253+43G>A, c.4539+2001G>A, or V4, c.4539+2064C>T).19,20,39 For all nine variants a splice effect was demonstrated via splice assays either in this study, by Sangermano et al. this issue (c.769–784C>T, c.4253+43G>A) or by Albert et al.26,38(c.4539+2001G>A). Six variants lead to PE inclusion into the transcript, all introducing a PTC; two variants induce (partial) exon skipping, leading to a frameshift and the introduction of a PTC; and lastly, one variant creates an in-frame insertion of three amino acids. Given the minor amount of aberrantly spliced transcript observed for the c.769–784C>T variant and its occurrence on the genotype c.[1454del](;)[5603A>T], its pathogenicity is currently considered unclear.
One of the spinouts of the identification of noncoding splicing variants is their potential to function as therapeutic targets for AON-mediated rescue. AON treatment for inherited diseases, including Duchenne muscular dystrophy, spinal muscular atrophy, and more recently Usher syndrome and Leber congenital amaurosis, has been or is being introduced in the clinic. Here, we used AON rescue to successfully correct the aberrant splicing induced by the variants c.859–540C>G, c.4539+1106C>T, and c.5197–557G>T in HEK 293-T cells and in patient-derived fibroblast cells carrying the c.4539+1106C>T variant. These results are of interest to patients, offering the potential to restore the amount of correct mRNA and subsequently of ABCA4 protein required for normal function in vivo.
Besides noncoding splice variants, we investigated the contribution and effect of putative cis-regulatory variants. Two noncoding deep-intronic variants (c.768+3223C>T and c.2919–383C>T) showed a significant downregulation using in vitro reporter studies in RPE-1 cells. It is unclear however if these assays in RPE-1 cells recapitulate the biological effect on ABCA4 expression in vivo. Even if the observed reduction is similar for both variants, the tested regulatory element in which c.768+3223C>T is located has a stronger basal expression and might thus have a larger effect on the overall ABCA4 expression.
Although ABCA4 variants represent one of the most prevalent causes of inherited retinal disease, its variant spectrum is characterized by a scarcity of structural variants (SVs), with only nine deletions and one complex rearrangement reported.3,7,10,11,12,13,14,15,16,17,18,40 Using a NGS CNV pipeline and high-resolution array-based CNV analysis, we identified six distinct CNVs in our cohort of monoallelic patients: four deletions, two of which are novel and two novel duplications. Interestingly, one of them is a noncoding tandem duplication located within the first intron of ABCA4 and encompassing several TFBSs. Delineation of the identified CNVs allowed an extensive bioinformatics analysis of their breakpoint regions revealing replicative-based mechanisms for all deletions and for the noncoding duplication, while the duplication spanning exons 2–6 most likely originated due to NHEJ. Next, we investigated the role of the hypomorphic c.5603A>T variant in the remaining cohort and identified it as a putative second allele in >72% of the remaining patients and >43% of the initial cohort, confirming its enrichment and clinical significance in monoallelic STGD1 patients, as recently described.23
Despite our approach, the molecular diagnosis remains uncertain in 11/67 (16.4%) patients. Some variants remain to be investigated via additional functional assays while other, more prevalent variants (MAF >1%) or splice variants without clear predictions have not been scrutinized in this study. Finally, it cannot be excluded that a subset of patients have pathogenic variants in other genes, representing phenocopies with a phenotype resembling ABCA4-associated disease.
To conclude, this study demonstrates that a locus-specific integrated approach combining genomics with downstream tailored functional studies is powerful for elucidating a major portion of missing heritability in ABCA4-associated disease. The discovery of novel pathogenic variants in noncoding regions and the development of AONs can be envisaged for personalized therapies. Overall, this ABCA4-oriented study can be regarded as a model for missing heritability in other autosomal recessive diseases with a recognizable phenotype and with an incomplete molecular diagnosis.
References
Allikmets R, Singh N, Sun H, et al. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet. 1997;15:236–246.
Cremers FP, van de Pol DJ, van Driel M, et al. Autosomal recessive retinitis pigmentosa and cone-rod dystrophy caused by splice site mutations in the Stargardt’s disease gene ABCR. Hum Mol Genet. 1998;7:355–362.
Lambertus S, van Huet RAC, Bax NM, et al. Early-onset Stargardt disease: phenotypic and genotypic characteristics. Ophthalmology. 2015;122:335–344.
Bertelsen M, Zernant J, Larsen M, Duno M, Allikmets R, Rosenberg T. Generalized choriocapillaris dystrophy, a distinct phenotype in the spectrum of ABCA4-associated retinopathies. Invest Ophthalmol Vis Sci. 2014;55:2766–2776.
Tanaka K, Lee W, Zernant J, et al. The rapid-onset chorioretinopathy phenotype of ABCA4 disease. Ophthalmology. 2018;125:89–99.
van Driel MA, Maugeri A, Klevering BJ, Hoyng CB, Cremers FP. ABCR unites what ophthalmologists divide(s). Ophthalmic Genet. 1998;19:117–122.
Maugeri A, van Driel MA, van de Pol DJ, et al. The 2588G-->C mutation in the ABCR gene is a mild frequent founder mutation in the Western European population and allows the classification of ABCR mutations in patients with Stargardt disease. Am J Hum Genet. 1999;64:1024–1035.
Cornelis SS, Bax NM, Zernant J, et al. In silico functional meta-analysis of 5,962 ABCA4 variants in 3,928 retinal dystrophy cases. Hum Mutat. 2017;38:400–408.
Sangermano R, Bax NM, Bauwens M, et al. Photoreceptor progenitor mRNA analysis reveals exon skipping resulting from the ABCA4 c.5461-10T→C mutation in Stargardt disease. Ophthalmology. 2016;123:1375–1385.
Bax NM, Sangermano R, Roosing S, et al. Heterozygous deep-intronic variants and deletions in ABCA4 in persons with retinal dystrophies and one exonic ABCA4 variant. Hum Mutat. 2015;36:43–47.
Yatsenko AN, Shroyer NF, Lewis RA, Lupski JR. An ABCA4 genomic deletion in patients with Stargardt disease. Hum Mutat. 2003;21:636–644.
Cideciyan AV, Swider M, Aleman TS, et al. Macular function in macular degenerations: repeatability of microperimetry as a potential outcome measure for ABCA4-associated retinopathy trials. Invest Ophthalmol Vis Sci. 2012;53:841–852.
Rozet JM, Gerber S, Ghazi I, et al. Mutations of the retinal specific ATP binding transporter gene (ABCR) in a single family segregating both autosomal recessive retinitis pigmentosa RP19 and Stargardt disease: evidence of clinical heterogeneity at this locus. J Med Genet. 1999;36:447–451.
Birch DG, Peters AY, Locke KL, Spencer R, Megarity CF, Travis GH. Visual function in patients with cone-rod dystrophy (CRD) associated with mutations in the ABCA4(ABCR) gene. Exp Eye Res. 2001;73:877–886.
Schorderet DF, Iouranova A, Favez T, Tiab L, Escher P. IROme, a new high-throughput molecular tool for the diagnosis of inherited retinal dystrophies. Biomed Res Int. 2013;2013:198089.
Lee W, Xie Y, Zernant J, et al. Complex inheritance of ABCA4 disease: four mutations in a family with multiple macular phenotypes. Hum Genet. 2016;135:9–19.
Carss KJ, Arno G, Erwood M, et al. Comprehensive rare variant analysis via whole-genome sequencing to determine the molecular pathology of inherited retinal disease. Am J Hum Genet. 2017;100:75–90.
Ellingford JM, Horn B, Campbell C, et al. Assessment of the incorporation of CNV surveillance into gene panel next-generation sequencing testing for inherited retinal diseases. J Med Genet. 2018;55:114–121.
Braun TA, Mullins RF, Wagner AH, et al. Non-exomic and synonymous variants in ABCA4 are an important cause of Stargardt disease. Hum Mol Genet. 2013;22:5136–5145.
Zernant J, Xie YA, Ayuso C, et al. Analysis of the ABCA4 genomic locus in Stargardt disease. Hum Mol Genet. 2014;23:6797–6806.
Bauwens M, De Zaeytijd J, Weisschuh N, et al. An augmented ABCA4 screen targeting noncoding regions reveals a deep intronic founder variant in Belgian Stargardt patients. Hum Mutat. 2015;36:39–42.
Schulz HL, Grassmann F, Kellner U, et al. Mutation spectrum of the ABCA4 gene in 335 Stargardt disease patients from a multicenter German cohort-impact of selected deep intronic variants and common SNPs. Invest Ophthalmol Vis Sci. 2017;58:394–403.
Zernant J, Lee W, Collison FT, et al. Frequent hypomorphic alleles account for a significant fraction of ABCA4 disease and distinguish it from age-related macular degeneration. J Med Genet. 2017;54:404–412.
Collin RW, Hollander den AI, van der Velde-Visser SD, Bennicelli J, Bennett J, Cremers FP. Antisense oligonucleotide (AON)-based therapy for Leber congenital amaurosis caused by a frequent mutation in CEP290. Mol Ther Nucleic Acids. 2012;1:e14.
Collin RWJ, Garanto A. Applications of antisense oligonucleotides for the treatment of inherited retinal diseases. Curr Opin Ophthalmol. 2017;28:260–266.
Albert S, Garanto A, Sangermano R. et al. Identification and rescue of splice defects caused by two neighboring deep-intronic ABCA4 mutations underlying Stargardt disease. Am J Hum Genet. 2018;102:517–527.
Slijkerman RW, Vaché C, Dona M, et al. Antisense oligonucleotide-based splice correction for USH2A-associated retinal degeneration caused by a frequent deep-intronic mutation. Mol Ther Nucleic Acids. 2016;5:e381.
Ahituv N, Prabhakar S, Poulin F, Rubin EM, Couronne O. Mapping cis-regulatory domains in the human genome using multi-species conservation of synteny. Hum Mol Genet. 2005;14:3057–3063.
Volders PJ, Verheggen K, Menschaert G, et al. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43:4363–4364.
Sangermano R, Khan M, Cornelis SS, et al. ABCA4 midigenes reveal the full splice spectrum of all reported noncanonical splice site variants in Stargardt disease. Genome Res. 2018;28:100–110.
Garanto A, Collin RWJ. Design and in vitro use of antisense oligonucleotides to correct pre-mRNA splicing defects in inherited retinal dystrophies. Methods Mol Biol. 2018;1715 suppl 1:61–78.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760.
DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498.
Plagnol V, Curtis J, Epstein M, et al. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics. 2012;28:2747–2754.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164–e164.
Van Cauwenbergh C, Van Schil K, Cannoodt R, et al. arrEYE: a customized platform for high-resolution copy number analysis of coding and noncoding regions of known and candidate retinal dystrophy genes and retinal noncoding RNAs. Genet Med. 2017;19:457–466.
Verdin H, D'haene B, Beysen D. et al. Microhomology-mediated mechanisms underlie non-recurrent disease-causing microdeletions of the FOXL2 gene or its regulatory domain. PLoS Genet. 2013;9:e1003358.
Sangermano R, Garanto A, Khan M, et al. Deep-intronic ABCA4 pathogenic variants explain missing heritability in Stargardt disease and allow correction of splice defects by antisense oligonucleotides. Genet Med. 2019;(in press). doi:10.1038/s41436-018.
Zernant J, Schubert C, Im KM, et al. Analysis of the Gene by Next-Generation Sequencing. Invest Ophthalmol Vis Sci. 2011;52:8479.
Falfoul Y, Habibi I, Turki A, Chebil A, Hassairi A, Schorderet DF, et al. Phenotypic progression of Stargardt disease in a large consanguineous Tunisian family harboring new ABCA4 mutations. Journal of Ophthalmology. 2018:1–7.
Acknowledgements
The authors gratefully acknowledge the families who participated in this study, Julie Van De Velde for technical assistance, and Silvia Albert for advice on cell culture studies. This study was supported by the Ghent University Special Research Fund (BOF15/GOA/011) to E.D.B., Hercules Foundation AUGE/13/023 to E.D.B., by the Research Foundation Flanders (G0C6715N and G0A9718N) to E.D.B., by the Foundation JED to E.D.B., and by Funds for Research in Ophthalmology (FRO) to M.B. S.N. is PhD fellow of the Institute for Innovation by Science and Technology (IWT), integrated in the FWO (FWO-SB). Further funds were provided by the FP7-PEOPLE-2012-ITN program EyeTN, agreement 317472 (to F.P.M.C.); the Foundation Fighting Blindness USA, grant number PPA-0517-0717-RAD (to A.G., F.P.M.C., and R.W.J.C.); the Macula Vision Research Foundation (to F.P.M.C.); the Rotterdamse Stichting Blindenbelangen, the Stichting Blindenhulp, and the Stichting tot Verbetering van het Lot der Blinden (to F.P.M.C.); and by the Landelijke Stichting voor Blinden en Slechtzienden, Macula Degeneratie fonds and the Stichting Blinden-Penning, which contributed through Uitzicht 2016-12 (to F.P.M.C.). This work was also supported by the Algemene Nederlandse Vereniging ter Voorkoming van Blindheid, Stichting Blinden-Penning, Landelijke Stichting voor Blinden en Slechtzienden, Stichting Oogfonds Nederland, Stichting Macula Degeneratie Fonds, and Stichting Retina Nederland Fonds, which contributed through UitZicht 2015-31, together with the Rotterdamse Stichting Blindenbelangen, Stichting Blindenhulp, Stichting tot Verbetering van het Lot der Blinden, Stichting voor Ooglijders, and Stichting Dowilvo (to A.G. and R.W.J.C.). This work was supported by the National Institute for Health Research England (NIHR) for the NIHR BioResource project (grant number RG65966). M.B. was PhD fellow of the FWO (11T9216N). E.D.B. (1802215N) and B.P.L. (1803816N) are senior clinical investigators of the FWO. The sponsors or funding organizations had no role in the design or conduct of this research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
The authors declare no conflicts of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, and provide a link to the Creative Commons license. You do not have permission under this license to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bauwens, M., Garanto, A., Sangermano, R. et al. ABCA4-associated disease as a model for missing heritability in autosomal recessive disorders: novel noncoding splice, cis-regulatory, structural, and recurrent hypomorphic variants. Genet Med 21, 1761–1771 (2019). https://doi.org/10.1038/s41436-018-0420-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41436-018-0420-y
Keywords
This article is cited by
-
Whole genome sequencing for inherited retinal diseases in the Korean National Project of Bio Big Data
Graefe's Archive for Clinical and Experimental Ophthalmology (2024)
-
Combining a prioritization strategy and functional studies nominates 5’UTR variants underlying inherited retinal disease
Genome Medicine (2024)
-
Introme accurately predicts the impact of coding and noncoding variants on gene splicing, with clinical applications
Genome Biology (2023)
-
Stargardt disease-associated in-frame ABCA4 exon 17 skipping results in significant ABCA4 function
Journal of Translational Medicine (2023)
-
Elusive variants in autosomal recessive disease: how can we improve timely diagnosis?
European Journal of Human Genetics (2023)