Abstract
Background
To explore the feasibility of artificial intelligence technology based on deep learning to automatically recognize the properties of vitreous opacities in ophthalmic ultrasound images.
Methods
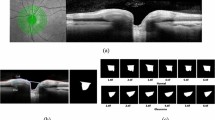
A total of 2000 greyscale Doppler ultrasound images containing non-pathological eye and three typical vitreous opacities confirmed as physiological vitreous opacity (VO), asteroid hyalosis (AH), and vitreous haemorrhage (VH) were selected and labelled for each lesion type. Five residual networks (ResNet) and two GoogLeNet models were trained to recognize vitreous lesions. Seventy-five percent of the images were randomly selected as the training set, and the remaining 25% were selected as the test set. The accuracy and parameters were recorded and compared among these seven different deep learning (DL) models. The precision, recall, F1 score, and area under the receiver operating characteristic curve (AUC) values for recognizing vitreous lesions were calculated for the most accurate DL model.
Results
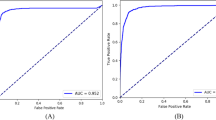
These seven DL models had significant differences in terms of their accuracy and parameters. GoogLeNet Inception V1 achieved the highest accuracy (95.5%) and minor parameters (10315580) in vitreous lesion recognition. GoogLeNet Inception V1 achieved precision values of 0.94, 0.94, 0.96, and 0.96, recall values of 0.94, 0.93, 0.97 and 0.98, and F1 scores of 0.94, 0.93, 0.96 and 0.97 for normal, VO, AH, and VH recognition, respectively. The AUC values for these four vitreous lesion types were 0.99, 1.0, 0.99, and 0.99, respectively.
Conclusions
GoogLeNet Inception V1 has shown promising results in ophthalmic ultrasound image recognition. With increasing ultrasound image data, a wide variety of confidential information on eye diseases can be detected automatically by artificial intelligence technology based on deep learning.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 18 print issues and online access
$259.00 per year
only $14.39 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Original data are in the possession of the corresponding author and are available on request.
References
Duke-Elder S. Diseases of the vitreous body. In: Henry Kimpton SD-E, editors. London. 1969; pp 315–75.
Baum G, Greenwood I. The application of ultrasonic locating techniques to ophthalmology; theoretic considerations and acoustic properties of ocular media. I. Reflective properties. Am J Ophthalmol. 1958;46:319–29.
Tandias R, Lemire CA, Palvadi K, Arroyo JG. Posterior vitreous detachment status a predictive factor for outcomes of vitrectomy for diabetic vitreous hemorrhage. Retina 2022;42:1103–10.
Lizzi FL, Coleman DJ. History of ophthalmic ultrasound. J Ultrasound Med. 2004;23:1255–66.
Liu SF, Wang Y, Yang X, Lei BY, Liu L, Li SX, et al. Deep learning in medical ultrasound analysis: a review. Engineering. 2019;5:261–75.
McCarthy J, Minsky ML, Rochester N, Shannon CE. A proposal for the dartmouth summer research project on artificial intelligence. AI MAG. 2006;27:12–4.
Samuel AL. Some studies in machine learning using the game of checkers. IBM J Res Dev. 2000;4:207–26.
Thrall JH, Li X, Li Q, Cruz C, Do S, Dreyer K, et al. Artificial intelligence and machine learning in radiology: opportunities, challenges, pitfalls, and criteria for success. J Am Coll Radio. 2018;15:504–8.
Shen D, Wu G, Suk HI. Deep learning in medical image analysis. Annu Rev Biomed Eng. 2017;19:221–48.
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88.
Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus Photographs. JAMA. 2016;316:2402–10.
van der Heijden AA, Abramoff MD, Verbraak F, van Hecke MV, Liem A, Nijpels G. Validation of automated screening for referable diabetic retinopathy with the IDx-DR device in the Hoorn Diabetes Care System. Acta Ophthalmol. 2018;96:63–8.
Burlina PM, Joshi N, Pekala M, Pacheco KD, Freund DE, Bressler NM. Automated grading of age-related macular degeneration from color fundus images using deep convolutional neural networks. JAMA Ophthalmol. 2017;135:1170–6.
Li Z, He Y, Keel S, Meng W, Chang RT, He M. Efficacy of a deep learning system for detecting glaucomatous optic neuropathy based on color fundus photographs. Ophthalmology. 2018;125:1199–206.
Varadarajan AV, Poplin R, Blumer K, Angermueller C, Ledsam J, Chopra R, et al. Deep learning for predicting refractive error from retinal fundus images. Invest Ophthalmol Vis Sci. 2018;59:2861–8.
Campbell JP, Ataer-Cansizoglu E, Bolon-Canedo V, Bozkurt A, Erdogmus D, Kalpathy-Cramer J, et al. Expert diagnosis of plus disease in retinopathy of prematurity from computer-based image analysis. JAMA Ophthalmol. 2016;134:651–7.
Poplin R, Varadarajan AV, Blumer K, Liu Y, McConnell MV, Corrado GS, et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng. 2018;2:158–64.
Moraru AD, Costin D, Moraru RL, Branisteanu DC. Artificial intelligence and deep learning in ophthalmology - present and future (Review). Exp Ther Med. 2020;20:3469–73.
Kermany DS, Goldbaum M, Cai W, Valentim CCS, Liang H, Baxter SL, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172:1122–31.
De Fauw J, Ledsam JR, Romera-Paredes B, Nikolov S, Tomasev N, Blackwell S, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018;24:1342–50.
Akkus Z, Cai J, Boonrod A, Zeinoddini A, Weston AD, Philbrick KA, et al. A Survey of deep-learning applications in ultrasound: artificial intelligence-powered ultrasound for improving clinical workflow. J Am Coll Radio. 2019;16:1318–28.
Wei W, Haishan X, Alpers J, Rak M, Hansen C. A deep learning approach for 2D ultrasound and 3D CT/MR image registration in liver tumor ablation. Comput Methods Prog Biomed. 2021;2016:106117.
Qian X, Pei J, Zheng H, Xie X, Yan L, Zhang H, et al. Prospective assessment of breast cancer risk from multimodal multiview ultrasound images via clinically applicable deep learning. Nat Biomed Eng. 2021;5:522–32.
Ossoinig K. Clinical echo- ophthalmology. In: Blodi FC, (ed). Current Concepts of Ophthalmology. 3. St Louis: CV Mosby Co; 1972. p. 101–30.
Tanter M, Fink M. Ultrafast imaging in biomedical ultrasound. IEEE Trans Ultrason Ferroelectr Freq Control. 2014;61:102–19.
Chen H, Zheng YF, Park JH, Heng PA, Zhou SK. Iterative multi-domain regularized deep learning for anatomical structure detection and segmentation from ultrasound images. Medical Image Computing and Computer-Assisted Intervention-MICCAI. 2016; 19th International Conference: 487–95.
Reddy UM, Filly RA, Copel JA. Prenatal imaging: ultrasonography and magnetic resonance imaging. Obstet Gynecol. 2008;112:145–57.
Rahimy E. Deep learning applications in ophthalmology. Curr Opin Ophthalmol. 2018;29:254–60.
Ting DSW, Peng L, Varadarajan AV, Keane PA, Burlina PM, Chiang MF, et al. Deep learning in ophthalmology: the technical and clinical considerations. Prog Retin Eye Res. 2019;72:100759.
Szegedy C, Liu W, Jia YQ, Sermanet P, Reed S, Anguelov D, et al. Going deeper with convolutions. Proceedings of the IEEE Comput Society Conference on Computer Vision and Pattern Recognition. 2015;7:1–9.
Laina I, Rupprecht C, Belagiannis V, Tombari F, Navab N. Deeper depth prediction with fully convolutional Residual networks. IEEE International Conference on 3D Vision. 2016;1–13.
Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. 32nd International Conference on machine learning. 2015;1:3.
He KM, Zhang XY, Ren SQ, Sun J. Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition. 2016;1:770–8.
Ma M, Gao Z, Wu J, Chen YL, Zheng X. A smile detection method based on improved LeNet-5 and support vector machine. 2018 IEEE SmartWorld/SCALCOM/UIC/ATC/CBDCom/IOP/SCI. 2018;446–51.
Chen D, Yu Y, Zhou Y, Peng B, Wang Y, Hu S, et al. A deep learning model for screening multiple abnormal findings in ophthalmic ultrasonography (With Video). Transl Vis Sci Technol. 2021;10:22.
Le C, Baroni M, Vinnett A, Levin MR, Martinez C, Jaafar M, et al. Deep learning model for accurate automatic determination of phakic status in pediatric and adult ultrasound biomicroscopy images. Transl Vis Sci Technol. 2020;9:63.
Shi G, Jiang Z, Deng G, Liu G, Zong Y, Jiang C, et al. Automatic classification of anterior chamber angle using ultrasound biomicroscopy and deep learning. Transl Vis Sci Technol. 2019;8:25.
Wang W, Wang L, Wang T, Wang X, Zhou S, Yang J, et al. Automatic localization of the scleral spur using deep learning and ultrasound biomicroscopy. Transl Vis Sci Technol. 2021;10:28.
Li W, Chen Q, Jiang Z, Deng G, Zong Y, Shi G, et al. Automatic anterior chamber angle measurement for ultrasound biomicroscopy using deep learning. J Glaucoma. 2020;29:81–5.
Thijssen JM. The history of ultrasound techniques in ophthalmology. Ultrasound Med Biol. 1993;19:599–618.
Lieb WE. Color Doppler imaging of the eye and orbit. Radio Clin North Am. 1998;36:1059–71.
Funding
This study was funded by Scientific Research Funding for Education Department of Liaoning Province (Grant No. ZD2020002).
Author information
Authors and Affiliations
Contributions
LF and MS were involved in the conception and conduct of the study, LF, HQ and MS were drafted this manuscript, and YZ, WW and MS were involved in review the manuscript. All authors read and approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Feng, L., Zhang, Y., Wei, W. et al. Applying deep learning to recognize the properties of vitreous opacity in ophthalmic ultrasound images. Eye 38, 380–385 (2024). https://doi.org/10.1038/s41433-023-02705-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41433-023-02705-7