Abstract
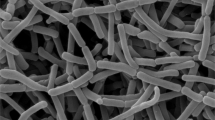
Strain Odt1-22T, an insect-derived actinomycete was isolated from a termite (Odontotermes formosanus) that was collected from Chanthaburi province, Thailand. Strain Odt1-22T was aerobic, Gram-stain-positive, and produced bud-like spore chain on the substrate hypha. According to chemotaxonomic analysis, strain Odt1-22T contained meso-diaminopimelic acid in peptidoglycan and the whole-cell hydrolysates contained arabinose, galactose, glucose, and ribose. The major menaquinone was MK-8(H4). The diagnostic phospholipids were diphosphatidylglycerol, hydroxyphosphatidylethanolamine, phosphatidylethanolamine and phosphatidylglycerol. Phylogenetic analysis based on 16 S rRNA gene sequence revealed that strain Odt1-22T was identified to the genus Actinomycetospora and showed high similarity values with A. chiangmaiensis DSM 45062 T (99.24%), A. soli SF1T (99.24%) and A. corticicola 014-5 T (98.17%). The genomic size of strain Odt1-22T was 6.6 Mbp with 73.8% G + C content and 6355 coding sequences (CDSs). The genomic analysis, strain Odt1-22T and closely related species A. chiangmaiensis DSM 45062 T, A. soli SF1T and A. corticicola DSM 45772 T displayed the values of average nucleotide identity-blast (ANIb) at 83.7–84.1% and MUMmer (ANIm) at 86.6–87.0%. Moreover, the results of digital DNA-DNA hybridization values between strain Odt1-22T and related Actinomycetospora species were 45.8−50.5% that lower than the threshold value of commonly used to delineate separated species level. On the basis of phenotypic, chemotaxonomic, and genotypic data, strain Odt1-22T represented a novel species within the genus Actinomycetospora, for which the name Actinomycetospora termitidis sp. nov. is proposed. The type strain of the species is Odt1-22T (= TBRC 16192 T = NBRC 115965 T).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Zhang Y, Liu C, Zhang J, Shen Y, Li C, He H. et al. Actinomycetospora atypica sp. nov., a novel soil actinomycete and emended description of the genus Actinomycetospora. Antonie van Leeuwenhoek. 2014;105:891–7.
Embley TM, O’Donnell AG, Rostron J, Goodfellow M. Chemotaxonomy of wall type IV actinomycetes which lack mycolic acids. Microbiology. 1988;134:953–60.
Jiang Y, Wiese J, Tang SK, Xu LH, Imhoff JF, Jiang CL. Actinomycetospora chiangmaiensis gen. nov., sp. nov., a new member of the family Pseudonocardiaceae. Int J Syst Evol Microbiol. 2008;58:408–413.
Yamamura H, Ashizawa H, Nakagawa Y, Hamada M, Ishida Y, Otoguro, M. et al. Actinomycetospora iriomotensis sp. nov., a novel actinomycete isolated from a lichen sample. J Antibiot. 2011;64:289–92.
Tamura T, Ishida Y, Hamada M, Otoguro M, Yamamura H, Hayakawa M. et al. Description of Actinomycetospora chibensis sp. nov., Actinomycetospora chlora sp. nov., Actinomycetospora cinnamomea sp. nov., Actinomycetospora corticicola sp. nov., Actinomycetospora lutea sp. nov., Actinomycetospora straminea sp. nov. and Actinomycetospora succinea sp. nov. and emended description of the genus Actinomycetospora. Int J Syst Evol Microbiol. 2011;61:1275–80.
Yamamura H, Ashizawa H, Nakagawa Y, Hamada M, Ishida Y, Otoguro M. et al. Actinomycetospora rishiriensis sp. nov., isolated from a lichen. Int J Syst Evol Microbiol. 2011;61:2621–2625.
He H, Zhang Y, Ma Z, Li C, Liu C, Zhou Y. et al. Actinomycetospora rhizophila sp. nov., an actinomycete isolated from rhizosphere soil of a peace lily (Spathi phyllum Kochii). Int J Syst Evol Microbiol. 2015;65:1520–4.
Sakdapetsiri C, Ngaemthao W, Suriyachadkun C, Duangmal K, Kitpreechavanich V. Actinomycetospora endophytica sp. nov., isolated from wild orchid (Podochilus microphyllus Lindl.) in Thailand. Int J Syst Evol Microbiol. 2018;68:3017–21.
Kaewkla C, Milton C, Franco M. Actinomycetospora callitridis sp. nov., an endophytic actinobacterium isolated from the surface-sterilised root of an Australian native pine tree. Antonie van Leeuwenhoek. 2019;112:331–7.
Chantavorakit T, Duangmal K. Actinomycetospora soli sp. nov., isolated from the rhizosphere soil of Averrhoa carambola L. Int J Syst Evol Microbiol. 2022;72:005277.
Hanshew AS, McDonald BR, Díaz CD, Djiéto-Lordon C, Blatrix R, Currie CR. et al. Characterization of actinobacteria associated with three ant-plant mutualisms. Micro Ecol. 2015;69:192–203.
Supong K, Anutrakunchai S, Thongkamngam T, Bunbamrung N, Pittayakhajonwut P, Tanasupawat S. et al. Investigation of biological activity and identification of culturable insect-derived Streptomyces strains from Cossus chlorates. Agr Nat Resour. 2023;57:271–280.
Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol 1966;16:313–40.
Kelly KL. Inter-society Color Council–National Bureau of standard color name charts illustrated with centroid colors. US Government Printing Office, Washington, DC, 1964.
Arai T. Culture media for actinomycetes. Tokyo: The Society for Actinomycetes, Japan. 1975.
Williams ST, Cross T. Actinomycetes. In Methods in Microbiology. Academic Press London, England. 1971;4:295–334.
Staneck JL, Roberts GD. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol. 1974;28:226–231.
Komagata K, Suzuki KI. Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol. 1987;19:161–207.
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A. et al. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods. 1984;2:233–241.
Sasser M. Identification of bacteria by gas chromatography of cellular fatty acids (MIDI Technical Note 101). Newark, DE: MIDI. 1990.
Kämpfer P, Kroppenstedt RM. Numerical analysis of fatty acid patterns of coryneform bacteria and related taxa. Can J Microbiol. 1996;42:989–1005.
Collins MD, Pirouz T, Goodfellow M, Minnikin DE. Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol. 1977;100:221–230.
Tamaoka J. Determination of DNA Base Composition. In: Goodfellow M, O’Donnell AG, editors. Chemical methods on prokaryotic systematics. Wiley, Chichester. 1994, pp 463–70.
Inahashi Y, Matsumoto A, Danbara H, Omura S, Takahashi Y. Phytohabitans suffuscus gen. nov., sp. nov., an actinomycete of the family Micromonosporaceae isolated from plant roots. Int J Syst Evol Microbiol. 2010;60:2652–2658.
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H. et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–7.
Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 1999;41:95–98.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25.
Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–76.
Kluge AG, Farris JS. Quantitative phyletics and the evolution of anurans. Syst Zool. 1969;18:1.
Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38:3022–3027.
Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20.
Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985;39:783–91.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77.
Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ. et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015;5:8365
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T. et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014;42:206–14.
Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10:2182.
Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016;32:929–31.
Meier-Kolthoff PJ, Alexander FA, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;14:60–73.
Lechevalier MP, Lechevalier HA. Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Bacteriol. 1970;20:435–43.
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91.
Kim M, Oh HS, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–51.
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kadler O, Krichevsky MI. et al. International committee on systematic bacteriology. Report of the ad hoc committee on the reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–4.
Acknowledgements
This work was financially supported by the Office of the Permanent Secretary, Ministry of Higher Education, Science Research, and Innovation (RGNS63-098).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Supong, K., Niemhom, N., Suriyachadkun, C. et al. Actinomycetospora termitidis sp. nov., an insect-derived actinomycete isolated from termite (Odontotermes formosanus). J Antibiot 77, 299–305 (2024). https://doi.org/10.1038/s41429-024-00712-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-024-00712-8