Abstract
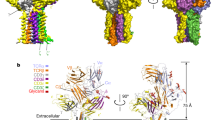
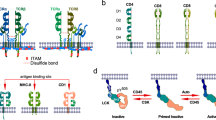
The T cell receptor (TCR) is one of the most complicated receptors in mammalian cells, and its triggering mechanism remains mysterious. As an octamer complex, TCR comprises an antigen-binding subunit (TCRαβ) and three CD3 signaling subunits (CD3ζζ, CD3δε, and CD3γε). Engagement of TCRαβ with an antigen peptide presented on the MHC leads to tyrosine phosphorylation of the immunoreceptor tyrosine-based activation motif (ITAM) in CD3 cytoplasmic domains (CDs), thus translating extracellular binding kinetics to intracellular signaling events. Whether conformational change plays an important role in the transmembrane signal transduction of TCR is under debate. Attracted by the complexity and functional importance of TCR, many groups have been studying TCR structure and triggering for decades using diverse biochemical and biophysical tools. Here, we synthesize these structural studies and discuss the relevance of the conformational change model in TCR triggering.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Smith-Garvin, J. E., Koretzky, G. A. & Jordan, M. S. T cell activation. Annu Rev. Immunol. 27, 591–619 (2009).
von Boehmer, H. & Kisielow, P. Self-nonself discrimination by T cells. Science 248, 1369–1373 (1990).
Pielak, R. M. et al. Early T cell receptor signals globally modulate ligand:receptor affinities during antigen discrimination. Proc. Natl Acad. Sci. USA 114, 12190–12195 (2017).
Huang, J. et al. A single peptide-major histocompatibility complex ligand triggers digital cytokine secretion in CD4(+) T cells. Immunity 39, 846–857 (2013).
Sykulev, Y., Joo, M., Vturina, I., Tsomides, T. J. & Eisen, H. N. Evidence that a single peptide-MHC complex on a target cell can elicit a cytolytic T cell response. Immunity 4, 565–571 (1996).
Schamel, W. W. et al. Coexistence of multivalent and monovalent TCRs explains high sensitivity and wide range of response. J. Exp. Med. 202, 493–503 (2005).
van der Merwe, P. A. & Dushek, O. Mechanisms for T cell receptor triggering. Nat. Rev. Immunol. 11, 47–55 (2011).
Courtney, A. H., Lo, W. L. & Weiss, A. TCR signaling: mechanisms of initiation and propagation. Trends Biochem. Sci. 43, 108–123 (2018).
Chakraborty, A. K. & Weiss, A. Insights into the initiation of TCR signaling. Nat. Immunol. 15, 798–807 (2014).
Hedrick, S. M., Cohen, D. I., Nielsen, E. A. & Davis, M. M. Isolation of cDNA clones encoding T cell-specific membrane-associated proteins. Nature 308, 149–153 (1984).
Yanagi, Y. et al. A human T cell-specific cDNA clone encodes a protein having extensive homology to immunoglobulin chains. Nature 308, 145–149 (1984).
Bentley, G. A., Boulot, G., Karjalainen, K. & Mariuzza, R. A. Crystal structure of the beta chain of a T cell antigen receptor. Science 267, 1984–1987 (1995).
Wang, J. et al. Atomic structure of an alphabeta T cell receptor (TCR) heterodimer in complex with an anti-TCR fab fragment derived from a mitogenic antibody. EMBO J. 17, 10–26 (1998).
Kjer-Nielsen, L. et al. Crystal structure of the human T cell receptor CD3 epsilon gamma heterodimer complexed to the therapeutic mAb OKT3. Proc. Natl Acad. Sci. USA 101, 7675–7680 (2004).
Arnett, K. L., Harrison, S. C. & Wiley, D. C. Crystal structure of a human CD3-epsilon/delta dimer in complex with a UCHT1 single-chain antibody fragment. Proc. Natl Acad. Sci. USA 101, 16268–16273 (2004).
Call, M. E. et al. The structure of the zetazeta transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell 127, 355–368 (2006).
Xu, C. et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-based motif. Cell 135, 702–713 (2008).
Garboczi, D. N. et al. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature 384, 134–141 (1996).
Yin, Y., Wang, X. X. & Mariuzza, R. A. Crystal structure of a complete ternary complex of T-cell receptor, peptide-MHC, and CD4. Proc. Natl Acad. Sci. USA 109, 5405–5410 (2012).
Garcia, K. C. et al. An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science 274, 209–219 (1996).
Rossjohn, J. et al. T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33, 169–200 (2015).
Dong De et al. Structural basis of assembly of the human T cell receptor-CD3 complex. Nature 573, 546–552 (2019).
Punt, J. A., Roberts, J. L., Kearse, K. P. & Singer, A. Stoichiometry of the T cell antigen receptor (TCR) complex: each TCR/CD3 complex contains one TCR alpha, one TCR beta, and two CD3 epsilon chains. J. Exp. Med. 180, 587–593 (1994).
Hennecke, J. & Wiley, D. C. T cell receptor-MHC interactions up close. Cell 104, 1–4 (2001).
Alcover, A., Alarcon, B. & Di Bartolo, V. Cell biology of T cell receptor expression and regulation. Annu. Rev. Immunol. 36, 103–125 (2018).
Ghendler, Y., Smolyar, A., Chang, H. C. & Reinherz, E. L. One of the CD3epsilon subunits within a T cell receptor complex lies in close proximity to the Cbeta FG loop. J. Exp. Med. 187, 1529–1536 (1998).
Kim, S. T. et al. Distinctive CD3 heterodimeric ectodomain topologies maximize antigen-triggered activation of alpha beta T cell receptors. J. Immunol. 185, 2951–2959 (2010).
Sasada, T. et al. Involvement of the TCR Cbeta FG loop in thymic selection and T cell function. J. Exp. Med. 195, 1419–1431 (2002).
Touma, M. et al. The TCR C beta FG loop regulates alpha beta T cell development. J. Immunol. 176, 6812–6823 (2006).
Das, D. K. et al. Pre-T cell receptors (Pre-TCRs) leverage vbeta complementarity determining regions (CDRs) and hydrophobic patch in mechanosensing thymic self-ligands. J. Biol. Chem. 291, 25292–25305 (2016).
Sun, Z. Y. et al. Solution structure of the CD3epsilondelta ectodomain and comparison with CD3epsilongamma as a basis for modeling T cell receptor topology and signaling. Proc. Natl Acad. Sci. USA 101, 16867–16872 (2004).
Birnbaum, M. E. et al. Molecular architecture of the alphabeta T cell receptor-CD3 complex. Proc. Natl Acad. Sci. USA 111, 17576–17581 (2014).
Sadelain, M., Riviere, I. & Riddell, S. Therapeutic T cell engineering. Nature 545, 423–431 (2017).
Backstrom, B. T., Muller, U., Hausmann, B. & Palmer, E. Positive selection through a motif in the alphabeta T cell receptor. Science 281, 835–838 (1998).
Werlen, G., Hausmann, B. & Palmer, E. A motif in the alphabeta T-cell receptor controls positive selection by modulating ERK activity. Nature 406, 422–426 (2000).
Backstrom, B. T. et al. A motif within the T cell receptor alpha chain constant region connecting peptide domain controls antigen responsiveness. Immunity 5, 437–447 (1996).
Mallaun, M. et al. The T cell receptor’s alpha-chain connecting peptide motif promotes close approximation of the CD8 coreceptor allowing efficient signal initiation. J. Immunol. 180, 8211–8221 (2008).
Naeher, D., Luescher, I. F. & Palmer, E. A role for the alpha-chain connecting peptide motif in mediating TCR-CD8 cooperation. J. Immunol. 169, 2964–2970 (2002).
Wang, Y. et al. A conserved CXXC motif in CD3epsilon is critical for T cell development and TCR signaling. PLoS Biol. 7, e1000253 (2009).
Xu, C., Call, M. E. & Wucherpfennig, K. W. A membrane-proximal tetracysteine motif contributes to assembly of CD3deltaepsilon and CD3gammaepsilon dimers with the T cell receptor. J. Biol. Chem. 281, 36977–36984 (2006).
Call, M. E., Pyrdol, J., Wiedmann, M. & Wucherpfennig, K. W. The organizing principle in the formation of the T cell receptor-CD3 complex. Cell 111, 967–979 (2002).
Geisler, C. Failure to synthesize the CD3-gamma chain. Consequences for T cell antigen receptor assembly, processing, and expression. J. Immunol. 148, 2437–2445 (1992).
Call, M. E. & Wucherpfennig, K. W. Molecular mechanisms for the assembly of the T cell receptor-CD3 complex. Mol. Immunol. 40, 1295–1305 (2004).
D’Oro, U. et al. Regulation of constitutive TCR internalization by the zeta-chain. J. Immunol. 169, 6269–6278 (2002).
Lauritsen, J. P. et al. Masking of the CD3 gamma di-leucine-based motif by zeta is required for efficient T-cell receptor expression. Traffic 5, 672–684 (2004).
Stenkamp, R. E., Teller, D. C. & Palczewski, K. Crystal structure of rhodopsin: a G-protein-coupled receptor. Chembiochem 3, 963–967 (2002).
Kuhns, M. S. & Davis, M. M. Disruption of extracellular interactions impairs T cell receptor-CD3 complex stability and signaling. Immunity 26, 357–369 (2007).
Kelly, L. A., Stephen, C. H., Wiley, D. C. Crystal structure of a human CD3ed dimer in complex with a UCHT1 single-chain antibody fragment. Proc. Natl Acad. Sci. USA 101, 16268–16273 (2004).
He, Y. et al. Identification of the docking site for CD3 on the T cell receptor beta chain by solution NMR. J. Biol. Chem. 290, 19796–19805 (2015).
Gao, G. F. et al. Crystal structure of the complex between human CD8alpha(alpha) and HLA-A2. Nature 387, 630–634 (1997).
Wang, R., Natarajan, K. & Margulies, D. H. Structural basis of the CD8 alpha beta/MHC class I interaction: focused recognition orients CD8 beta to a T cell proximal position. J. Immunol. 183, 2554–2564 (2009).
Krshnan, L., Park, S., Im, W., Call, M. J. & Call, M. E. A conserved alphabeta transmembrane interface forms the core of a compact T-cell receptor-CD3 structure within the membrane. Proc. Natl Acad. Sci. USA 113, E6649–E6658 (2016).
Swamy, M. et al. A cholesterol-based allostery model of T cell receptor phosphorylation. Immunity 44, 1091–1101 (2016).
Matsui, K. et al. Low affinity interaction of peptide-MHC complexes with T cell receptors. Science 254, 1788–1791 (1991).
Gee, M. H. et al. Stress-testing the relationship between T cell receptor/peptide-MHC affinity and cross-reactivity using peptide velcro. Proc. Natl Acad. Sci. USA 115, E7369–E7378 (2018).
Yin, Y. & Mariuzza, R. A. The multiple mechanisms of T cell receptor cross-reactivity. Immunity 31, 849–851 (2009).
Birnbaum, M. E. et al. Deconstructing the peptide-MHC specificity of T cell recognition. Cell 157, 1073–1087 (2014).
Wu, L. C., Tuot, D. S., Lyons, D. S., Garcia, K. C. & Davis, M. M. Two-step binding mechanism for T-cell receptor recognition of peptide MHC. Nature 418, 552–556 (2002).
Artyomov, M. N., Lis, M., Devadas, S., Davis, M. M. & Chakraborty, A. K. CD4 and CD8 binding to MHC molecules primarily acts to enhance Lck delivery. Proc. Natl Acad. Sci. USA 107, 16916–16921 (2010).
McKeithan, T. W. Kinetic proofreading in T-cell receptor signal transduction. Proc. Natl Acad. Sci. USA 92, 5042–5046 (1995).
Thill, P. A., Weiss, A. & Chakraborty, A. K. Phosphorylation of a tyrosine residue on Zap70 by Lck and its subsequent binding via an SH2 domain may be a key gatekeeper of T cell receptor signaling in vivo. Mol. Cell Biol. 36, 2396–2402 (2016).
Yousefi O. S., et al. Optogenetic control shows that kinetic proofreading regulates the activity of the T cell receptor. Elife 8, 42475 (2019).
Govern, C. C., Paczosa, M. K., Chakraborty, A. K. & Huseby, E. S. Fast on-rates allow short dwell time ligands to activate T cells. Proc. Natl Acad. Sci. USA 107, 8724–8729 (2010).
Aleksic, M. et al. Dependence of T cell antigen recognition on T cell receptor-peptide MHC confinement time. Immunity 32, 163–174 (2010).
Stepanek, O. et al. Coreceptor scanning by the T cell receptor provides a mechanism for T cell tolerance. Cell 159, 333–345 (2014).
Valitutti, S. The serial engagement model 17 years after: from TCR triggering to immunotherapy. Front. Immunol. 3, 272 (2012).
Valitutti, S., Muller, S., Cella, M., Padovan, E. & Lanzavecchia, A. Serial triggering of many T-cell receptors by a few peptide-MHC complexes. Nature 375, 148–151 (1995).
Yang, W. et al. Dynamic regulation of CD28 conformation and signaling by charged lipids and ions. Nat. Struct. Mol. Biol. 24, 1081–1092 (2017).
Das, J. et al. Digital signaling and hysteresis characterize ras activation in lymphoid cells. Cell 136, 337–351 (2009).
Stefanova, I. et al. TCR ligand discrimination is enforced by competing ERK positive and SHP-1 negative feedback pathways. Nat. Immunol. 4, 248–254 (2003).
Wertek, F. & Xu, C. Digital response in T cells: to be or not to be. Cell Res. 24, 265–266 (2014).
Hermiston, M. L., Xu, Z. & Weiss, A. CD45: a critical regulator of signaling thresholds in immune cells. Annu. Rev. Immunol. 21, 107–137 (2003).
Chang, V. T. et al. Initiation of T cell signaling by CD45 segregation at ‘close contacts’. Nat. Immunol. 17, 574–582 (2016).
Choudhuri, K., Wiseman, D., Brown, M. H., Gould, K. & van der Merwe, P. A. T-cell receptor triggering is critically dependent on the dimensions of its peptide-MHC ligand. Nature 436, 578–582 (2005).
Courtney, A. H. et al. A Phosphosite within the SH2 domain of Lck regulates its activation by CD45. Mol. Cell 67, 498–511 (2017).
Janeway, C. A. Jr. Ligands for the T-cell receptor: hard times for avidity models. Immunol. Today 16, 223–225 (1995).
Liu, B. et al. 2D TCR-pMHC-CD8 kinetics determines T-cell responses in a self-antigen-specific TCR system. Eur. J. Immunol. 44, 239–250 (2014).
Huang, J. et al. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature 464, 932–936 (2010).
Hawse, W. F. et al. TCR scanning of peptide/MHC through complementary matching of receptor and ligand molecular flexibility. J. Immunol. 192, 2885–2891 (2014).
Krogsgaard, M. et al. Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T cell activation. Mol. Cell 12, 1367–1378 (2003).
Schon, A. & Freire, E. Thermodynamics of intersubunit interactions in cholera toxin upon binding to the oligosaccharide portion of its cell surface receptor, ganglioside GM1. Biochemistry 28, 5019–5024 (1989).
Garcia, K. C. et al. Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen. Science 279, 1166–1172 (1998).
Holland, C. J. et al. In silico and structural analyses demonstrate that intrinsic protein motions guide T cell receptor complementarity determining region loop flexibility. Front. Immunol. 9, 674 (2018).
Reiser, J. B. et al. CDR3 loop flexibility contributes to the degeneracy of TCR recognition. Nat. Immunol. 4, 241–247 (2003).
Rudolph, M. G., Stanfield, R. L. & Wilson, I. A. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24, 419–466 (2006).
Beddoe, T. et al. Antigen ligation triggers a conformational change within the constant domain of the alphabeta T cell receptor. Immunity 30, 777–788 (2009).
Rangarajan, S. et al. Peptide-MHC (pMHC) binding to a human antiviral T cell receptor induces long-range allosteric communication between pMHC- and CD3-binding sites. J. Biol. Chem. 293, 15991–16005 (2018).
Reiser, J. B. et al. A T cell receptor CDR3beta loop undergoes conformational changes of unprecedented magnitude upon binding to a peptide/MHC class I complex. Immunity 16, 345–354 (2002).
Natarajan, K. et al. An allosteric site in the T-cell receptor Cbeta domain plays a critical signalling role. Nat. Commun. 8, 15260 (2017).
Chen, Y., Ju, L., Rushdi, M., Ge, C. & Zhu, C. Receptor-mediated cell mechanosensing. Mol. Biol. Cell 28, 3134–3155 (2017).
Anvari, B., Torres, J. H. & McIntyre, B. W. Regulation of pseudopodia localization in lymphocytes through application of mechanical forces by optical tweezers. J. Biomed. Opt. 9, 865–872 (2004).
Basu, R. et al. Cytotoxic T cells use mechanical force to potentiate target cell killing. Cell 165, 100–110 (2016).
Hong, J. et al. Force-regulated in situ TCR–peptide-bound MHC class II kinetics determine functions of CD4+ T cells. J. Immunol. 195, 3557–3564 (2015).
Kochenderfer, J. N. et al. Eradication of B-lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood 116, 4099 (2010).
Kim, S. T. et al. The alphabeta T cell receptor is an anisotropic mechanosensor. J. Biol. Chem. 284, 31028–31037 (2009).
Adams, J. J. et al. T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex. Immunity 35, 681–693 (2011).
Feng, D., Bond, C. J., Ely, L. K., Maynard, J. & Garcia, K. C. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat. Immunol. 8, 975–983 (2007).
Marshall, B. T. et al. Direct observation of catch bonds involving cell-adhesion molecules. Nature 423, 190–193 (2003).
Suzuki, T. et al. Mechanical force effect on the two-state equilibrium of the hyaluronan-binding domain of CD44 in cell rolling. Proc. Natl Acad. Sci. USA 112, 6991–6996 (2015).
Sibener, L. V. et al. Isolation of a structural mechanism for uncoupling T cell receptor signaling from peptide-MHC binding. Cell 174, 672–687 (2018).
Liu, Y. et al. DNA-based nanoparticle tension sensors reveal that T-cell receptors transmit defined pN forces to their antigens for enhanced fidelity. Proc. Natl Acad. Sci. USA 113, 5610–5615 (2016).
Das, D. K. et al. Force-dependent transition in the T-cell receptor beta-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc. Natl Acad. Sci. USA 112, 1517–1522 (2015).
Pryshchep, S., Zarnitsyna, V. I., Hong, J., Evavold, B. D. & Zhu, C. Accumulation of serial forces on TCR and CD8 frequently applied by agonist antigenic peptides embedded in MHC molecules triggers calcium in T cells. J. Immunol. 193, 68–76 (2014).
Liu, B., Chen, W., Evavold, B. D. & Zhu, C. Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell 157, 357–368 (2014).
Wu, P. et al. Mechano-regulation of peptide-MHC class I conformations determines TCR antigen recognition. Mol. Cell 73, 1015–1027 (2019).
Wieczorek, M. et al. Major histocompatibility complex (MHC) class I and MHC class II proteins: conformational plasticity in antigen presentation. Front. Immunol. 8, 292 (2017).
Fernandes, R. A. et al. T cell receptors are structures capable of initiating signaling in the absence of large conformational rearrangements. J. Biol. Chem. 287, 13324–13335 (2012).
Lee, M. S. et al. A mechanical switch couples T cell receptor triggering to the cytoplasmic juxtamembrane regions of CD3zetazeta. Immunity 43, 227–239 (2015).
Brazin, K. N. et al. The T cell antigen receptor alpha transmembrane domain coordinates triggering through regulation of bilayer immersion and CD3 subunit associations. Immunity 49, 829–841 (2018).
Schamel, W. W., Alarcon, B., Hofer, T. & Minguet, S. The allostery model of TCR regulation. J. Immunol. 198, 47–52 (2017).
Beck-Garcia, K. et al. Nanoclusters of the resting T cell antigen receptor (TCR) localize to non-raft domains. Biochim. Biophys. Acta 1853, 802–809 (2015).
Yang, W. et al. Potentiating the antitumour response of CD8(+) T cells by modulating cholesterol metabolism. Nature 531, 651–655 (2016).
Wang, F., Beck-Garcia, K., Zorzin, C., Schamel, W. W. & Davis, M. M. Inhibition of T cell receptor signaling by cholesterol sulfate, a naturally occurring derivative of membrane cholesterol. Nat. Immunol. 17, 844–850 (2016).
Nika, K. et al. Constitutively active Lck kinase in T cells drives antigen receptor signal transduction. Immunity 32, 766–777 (2010).
Ballek, O., Valecka, J., Manning, J. & Filipp, D. The pool of preactivated Lck in the initiation of T-cell signaling: a critical re-evaluation of the Lck standby model. Immunol. Cell Biol. 93, 384–395 (2015).
Aivazian, D. & Stern, L. J. Phosphorylation of T cell receptor zeta is regulated by a lipid dependent folding transition. Nat. Struct. Biol. 7, 1023–1026 (2000).
Deford-Watts, L. M. et al. The cytoplasmic tail of the T cell receptor CD3 epsilon subunit contains a phospholipid-binding motif that regulates T cell functions. J. Immunol. 183, 1055–1064 (2009).
Zhang, H., Cordoba, S. P., Dushek, O. & van der Merwe, P. A. Basic residues in the T-cell receptor zeta cytoplasmic domain mediate membrane association and modulate signaling. Proc. Natl Acad. Sci. USA 108, 19323–19328 (2011).
DeFord-Watts, L. M. et al. The CD3 zeta subunit contains a phosphoinositide-binding motif that is required for the stable accumulation of TCR-CD3 complex at the immunological synapse. J. Immunol. 186, 6839–6847 (2011).
Bettini, M. L. et al. Membrane association of the CD3epsilon signaling domain is required for optimal T cell development and function. J. Immunol. 193, 258–267 (2014).
Li, L. et al. Ionic CD3-Lck interaction regulates the initiation of T-cell receptor signaling. Proc. Natl Acad. Sci. USA 114, E5891–E5899 (2017).
Guo, X. et al. Lipid-dependent conformational dynamics underlie the functional versatility of T-cell receptor. Cell Res. 27, 505–525 (2017).
Shi, X. et al. Ca2+ regulates T-cell receptor activation by modulating the charge property of lipids. Nature 493, 111–115 (2013).
Sasmal, D. K. et al. TCR-pMHC bond conformation controls TCR ligand discrimination. Cell. Mol. Immunol. (2019). https://doi.org/10.1038/s41423-019-0273-6. [Epub ahead of print].
Dobbins, J. et al. Binding of the cytoplasmic domain of CD28 to the plasma membrane inhibits Lck recruitment and signaling. Sci. Signal. 9, ra75 (2016).
Gil, D., Schamel, W. W., Montoya, M., Sanchez-Madrid, F. & Alarcon, B. Recruitment of Nck by CD3 epsilon reveals a ligand-induced conformational change essential for T cell receptor signaling and synapse formation. Cell 109, 901–912 (2002).
Paensuwan, P. et al. Nck binds to the T cell antigen receptor using its SH3.1 and SH2 domains in a cooperative manner, promoting TCR functioning. J. Immunol. 196, 448–458 (2016).
Hem, C. D. et al. T cell specific adaptor protein (TSAd) promotes interaction of Nck with Lck and SLP-76 in T cells. Cell Commun. Signal. 13, 31 (2015).
Borroto, A. et al. First-in-class inhibitor of the T cell receptor for the treatment of autoimmune diseases. Sci. Transl. Med. 8, 370ra184 (2016).
Sun, Z. J., Kim, K. S., Wagner, G. & Reinherz, E. L. Mechanisms contributing to T cell receptor signaling and assembly revealed by the solution structure of an ectodomain fragment of the CD3 epsilon gamma heterodimer. Cell 105, 913–923 (2001).
Ding, Y. H., Baker, B. M., Garboczi, D. N., Biddison, W. E. & Wiley, D. C. Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical. Immunity 11, 45–56 (1999).
Kjer-Nielsen, L. et al. The 1.5 A crystal structure of a highly selected antiviral T cell receptor provides evidence for a structural basis of immunodominance. Structure 10, 1521–1532 (2002).
Kjer-Nielsen, L. et al. A structural basis for the selection of dominant alphabeta T cell receptors in antiviral immunity. Immunity 18, 53–64 (2003).
Acknowledgements
We thank Wei Wu and Chengsong Yan for thoughtful discussions. C.X. is funded by CAS grants (Strategic Priority Research Program XDB29000000, Facility-based Open Research Program QYZDB-SSW-SMC048, Fountain-Valley Life Sciences Fund of University of Chinese Academy of Sciences Education Foundation), NSFC grant (31861133009, 31621003), MOST Grant (2018YFA0800700) and the Ten Thousand Talent Program “Leading Talent” of China. H.L. is funded by an NSFC grant (31670751).
Author information
Authors and Affiliations
Contributions
C.X. designed the framework. X.X. wrote the manuscript. H.L. and C.X. revised it. H.L. and X.X. made the figures.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Xu, X., Li, H. & Xu, C. Structural understanding of T cell receptor triggering. Cell Mol Immunol 17, 193–202 (2020). https://doi.org/10.1038/s41423-020-0367-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-020-0367-1
Keywords
This article is cited by
-
The pathogen-encoded signalling receptor Tir exploits host-like intrinsic disorder for infection
Communications Biology (2024)
-
Membrane-anchored DNA nanojunctions enable closer antigen-presenting cell–T-cell contact in elevated T-cell receptor triggering
Nature Nanotechnology (2023)
-
TREM2/PLCγ2 signalling in immune cells: function, structural insight, and potential therapeutic modulation
Molecular Neurodegeneration (2021)
-
T cell receptor (TCR) signaling in health and disease
Signal Transduction and Targeted Therapy (2021)