Abstract
The tumor suppressor p53 is usually inactivated by somatic mutations in malignant neoplasms, and its reactivation represents an attractive therapeutic strategy for cancers. Here, we reported that a new quinolone compound RYL-687 significantly inhibited non-small cell lung cancer (NSCLC) cells which express wild type (wt) p53, in contract to its much weaker cytotoxicity on cells with mutant p53. RYL-687 upregulated p53 in cells with wt but not mutant p53, and ectopic expression of wt p53 significantly enhanced the anti-NSCLC activity of this compound. RYL-687 induced production of reactive oxygen species (ROS) and upregulation of Nrf2, leading to an elevation of the NAD(P)H:quinoneoxidoreductase-1 (NQO1) that can protect p53 by inhibiting its degradation by 20S proteasome. RYL-687 bound NQO1, facilitating the physical interaction between NQO1 and p53. NQO1 was required for RYL-687-induced p53 accumulation, because silencing of NQO1 by specific siRNA or an NQO1 inhibitor uridine, drastically suppressed RYL-687-induced p53 upregulation. Moreover, a RYL-687-related prodrug significantly inhibited tumor growth in NOD-SCID mice inoculated with NSCLC cells and in a wt p53-NSCLC patient-derived xenograft mouse model. These data indicate that targeting NQO1 is a rational strategy to reactivate p53, and RYL-687 as a p53 stabilizer bears therapeutic potentials in NSCLCs with wt p53.
Similar content being viewed by others
Introduction
Lung cancer, the most common cause of cancer-related mortality worldwide [1], can be divided into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC); the latter type consists of lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and large cell carcinoma [2]. In the past 20 years, the use of targeted therapies and immunotherapy has achieved survival benefits in a certain proportion of patients [3, 4]. However, the 5-year overall survival of NSCLCs each stage combined was only 18.6% in 2019 [5, 6]; therefore, there is an urgent need to find novel therapeutic targets to develop new treatment approaches to improve patient outcome.
The quinolone drug family is a very important family of antibiotics and is used as a broad-spectrum antibiotic with a low rate of severe adverse effects. The quinolone family is one of the most important drugs in the pharmaceutical chemistry. Quinolone drugs also have anticancer [7,8,9] and anti-inflammatory biological activities [10]. Quinolones such as doxorubicin, enoxacin, etoposide and irinotecan, have been widely used as anti-tumor drugs [11]. There are an increasing number of reports that quinolones can induce apoptosis and growth inhibition in various tumor cells [12,13,14]. Some quinolone drugs can reverse drug resistance of cancer cells [15], and some others are being tested in different stages of clinical trials [16, 17].
Previously, our chemical team exploited a library by phenotypic screening to discover new antiviral inhibitors [18]. We identified a novel small compound RYL-634, which is a quinolone compound, shows excellent broad-spectrum inhibitory activity against various viruses including hepatitis C virus, dengue virus, Zika virus, human immunodeficiency virus, and others [18]. Based on the structure of RYL-634, a series of analogues including compound RYL-687 (Supplementary Table 1) were optimally designed and synthesized. In this study, we tested the effects of these analogues on NSCLC cells and investigated the effect of RYL-687 on the expression of p53. We reported that RYL-687 could stabilize wild type (wt) p53 by targeting the NAD(P)H:quinone-oxidoreductase-1 (NQO1) that is a two-electron reductase responsible for detoxification of quinones and represents an emerging target for anticancer drug development [19].
Materials and methods
Antibodies and reagents
Antibodies used included rabbit anti-human NQO1 (#62262S, Cell Signaling Technology; 1:1000 for Western blot; 1:200 for immunofluorescence), mouse anti-human p53 (#2524, Cell Signaling Technology; 1:1000 for Western blot; 1:2000 for immunofluorescence; 1:500 for immunoprecipitation), rabbit anti-human MDM2 (#86934S, Cell Signaling Technology; 1:1000 for Western blot), rabbit anti-human p21 (#2947T, rabbit anti-human, Cell Signaling Technology; 1:1000 for Western blot), mouse anti-human β-Actin (#A5441, Sigma-Aldrich; 1:5000 for Western blot), rabbit anti-human Bax (#2947T, Cell Signaling Technology; 1:1000 for Western blot), rabbit anti-human PARP (# 9542S, Cell Signaling Technology; 1:1000 for Western blot), rabbit anti-human BCL2 (#2827S, Cell Signaling Technology; 1:1000 for Western blot), rabbit anti-human Cleaved-Casp 3 (#9664S, Cell Signaling Technology; 1:1000 for Western blot), rabbit anti-human Cleaved-Casp 9 (#9505, Cell Signaling Technology; 1:1000 for Western blot), mouse anti-human α-Tubulin (#sc5286, Santa Cruz Biotechnology; 1:500 for Western blot), rabbit anti-human CDK1 (#ab201008, Abcam; 1:1000 for Western blot), rabbit anti-human CDK2 (#18048, Cell Signaling Technology; 1:1000 for Western blot), rabbit anti-human CDK4 (#12790, Cell Signaling Technology; 1:1000 for Western blot), rabbit anti-human Cyclin A2 (#A19036, ABclonal; 1:1000 for Western blot), mouse anti-human Cyclin B1 (#4135, Cell Signaling Technology; 1:1000 for Western blot), mouse anti-human Cyclin D1 (#2926, Cell Signaling Technology; 1:1000 for Western blot), rabbit anti-human Cyclin E1 (#20808, Cell Signaling Technology; 1:1000 for Western blot), rabbit anti-human Nrf2 (#12721, Cell Signaling Technology; 1:1000 for Western blot), rabbit anti-human γ-H2AX (# 2577S, Cell Signaling Technology; 1:1000 for Western blot), goat anti-human Lamin B (#sc6216, Santa Cruz Biotechnology; 1:1000 for Western blot), rabbit anti-human cytochrome c (#A13430, ABclonal; 1:1000 for Western blot). Quinolone compound RYL-634 and 5 analogues (Supplementary Table 1) were synthesized by our chemical team; taxol (#S1150) and AZD9291 (#S7297) were obtained from Selleckchem China (Shanghai, China).
Cell culture
The human NSCLC cell lines A549, H1299, H1975, H460, HOP-62, HCC827 and H520 were was obtained from the American Type Culture Collection [ATCC] (Manassas, VA, USA). The cells were cultured in medium DMEM (A549) or RPMI-1640 (H460, H1975, H1299, HOP-62, HCC827 and H520) supplemented with 10% foetal bovine serum (FBS; Gibco/BRL, Grand Island, NY, USA).
Cell proliferation, cell apoptosis, and colony formation activity assays
The cells were seeded onto 96-well plates and pre-cultured for 24 h, treated with indicated compounds for 24–72 h, and co-incubated with CellTiter 96®AQueous One Solution cell proliferation assay reagents (Promega, Madison, WI, USA) that contain 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS), and cell proliferation was analyzed according to manufacturer’s instruction. The absorbance at 490 nm was measured using a microplate reader (Bio-Tek, Winooski, VT, USA) and the concentration causing 50% cell growth inhibition (GI50) was calculated. Cell cycle and cell apoptosis were respectively evaluated by propidium iodide (PI)-staining of cold ethanol fixed cells and the Annexin V-FITC Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA), followed by analyses using a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA). For colony formation assay, the cells were seeded onto 6-well plates and treated with RYL-687 at indicated concentrations. After 10 days of culturing, the medium was removed and 1 mL of methanol was added to each well to fix the cells for 30 min at room temperature. The cells were stained with 0.005% crystal violet (Sigma, St. Louis, MO, USA) for 30 min, washed with PBS until the background color was removed, and colonies containing 50 cells or more were counted.
Small interfering RNAs (siRNAs), plasmids, single-guide RNAs (sgRNAs), and transfection
The sequences of siRNAs and sgRNAs were listed in Supplementary Table 1. The lentiCRISPR-Cas9 plasmids were obtained from Addgene (Watertown, MA, USA), and sgRNAs were purchased from Tsingke Biotechnology (Beijing, China). The cells were transfected with siRNAs or pcDNA3.0 plasmids (Invitrogen) containing wt TP53 using Lipofectamine 3000 (Invitrogen, California, CA, USA) according to the manufacturer’s instructions. The lentiCRISPR plasmids and sgRNAs were transfected into H1975 cells as described [20], puromycin resistance cells were selected, and the gene knockout events were verified by sequencing analysis of the genomic PCR fragments of the target loci. The cells were then transfected with pcDNA3.0-wt TP53 plasmids, treated with RYL-687 for 48 h, and co-incubated with CellTiter 96®AQueous One Solution to analyze cell viability.
Identification of RYL-687 direct targets
A biotin-labeled RYL-687 (Bio-687, Supplementary Fig. 1a) was synthesized to identify the direct targets of the compound. Cells upon Bio-687 were lysed with lysis buffer, cell lysates were incubated with streptavidin agarose overnight at 4 °C. After washing with lysis buffer, streptavidin agarose beads were resuspended in SDS loading buffer [21]. The proteins were electrophoresed, and bands of interest were analyzed by mass spectrum.
Measurement of intracellular reactive oxygen species
The levels of reactive oxygen species (ROS) in H460 cells were determined by using the 2,7-dichlorofluorescin diacetate (DCFHDA) assay. The cells were treated with RYL-687 at indicated concentration for 24 h, washed with PBS, and co-incubated with 10 μM for 30 min at 37 °C. The cells were then washed with PBS and analysed by a flow cytometer.
Immunofluorescence analysis
H460 and A549 cells were plated onto glass slides with 1% gelatine in 6-well plates, treated with RYL-687 at indicated protocols, washed twice with PBS, and fixed with 4% paraformaldehyde for 30 min at room temperature. The cells were washed with 150 mM glycine in PBS and permeabilized with 0.1% Triton X-100 in PBS for 20 min. After washing three times with 0.05% Tween in PBS, cell smears were blocked with 5% BSA for 1 h at room temperature. Then, the cell smears were incubated with the indicated primary antibodies at 4 °C overnight, washed, and incubated with FITC-conjugated secondary antibody for 2 h at room temperature. The nuclei were stained with DAPI. Images were taken by a laser confocal scanning microscope (N-STORM, Nikon, Tokyo, Japan).
Western blot analysis
The cells were harvested, washed with PBS, and lysed on ice for 30 min in RIPA buffer supplemented with the protease inhibitors cocktail (Sigma, St Louis, MO), and protein extracts were quantitated. Equal amounts of lysates were separated by 10%–15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (Millipore Corporation, Darmstadt, Germany), which were blocked with 5% nonfat milk in Tris-buffered saline for 1 h and washed. After incubation with the indicated primary and secondary antibodies, the signal on the membranes was detected using a Luminescent Image Analyser LSA 4000 (GE, Fairfield, CO, USA).
Animal studies
All animal studies were conducted according to protocols approved by the Animal Ethics Committee of the Tsinghua University School of Medicine. The NOD-SCID mice were bred and maintained in a specific pathogen-free environment. The mice were inoculated with H460 (1 × 106) cells, and were treated with vehicle control (5% DMSO, 10% Castor Oil, 85% PBS) or RYL-687 related quinolone prodrug (hereafter, Prodrug; Supplementary Table 1) when the tumors reached a palpable size. The reason for the use of the Prodrug was the poor water solubility of RYL-687. The Prodrug was intraperitoneally administered at 20 and 60 mg·kg−1·d−1, once per day for 3 weeks. Chemotherapeutic agent methotrexate, which was used as a positive control, was intraperitoneally administered at a dosage of 2 mg/kg, once per day for 3 weeks. Caliper measurements of the longest perpendicular tumor diameters were performed every other day to estimate the tumor volume, using the following formula: 4π/3 × (width/2)2 × (length/2), representing the 3-dimensional volume of an ellipse. Animals were sacrificed when tumors reached 2 cm or if the mice appeared moribund to prevent unnecessary morbidity to the mice.
Statistical analysis
All experiments were repeated at least three times, and the data are presented as the mean ± SD unless noted otherwise. Statistical analyses were conducted using GraphPad Prism 5 (GraphPad Software, La Jolla, CA). Statistically significant differences were determined by Student’s t test or Mann–Whitney test. P values less than 0.05 indicate statistical significance.
Results
RYL-687 exhibits potent inhibitory effects on NSCLC cells
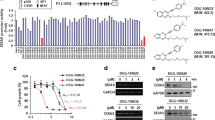
Our chemical team constructed a library of biaryl-substituted quinolones through phenotypic screening and identified a new antiviral compound, RYL-634, which demonstrated excellent broad-spectrum inhibition activity against various viruses [18]. It was well known that quinolones bear many biological activities, while biaryl-substituted quinolones had rarely been explored for their antitumor effects [22]. Here, we investigated the effects of RYL-634 and its 4 derivatives (Supplementary Table 1) on 4 NSCLC lines (H460, A549, H1975 and H1299) to identify the best candidate for the following experiments. Taxol and AZD9291 were used as positive controls. The results showed that these compounds could significantly inhibit lung cancer cell growth (Fig. 1a), while RYL-687 (Fig. 1b) demonstrated the strongest inhibitory effect among these RYL-634-derived molecules, with GI50 (72 h) values of 22.29, 23.19, 83.17, and >10000 nM in the H460, A549, H1975, and H1299 cell lines, respectively (Fig. 1a).
a NSCLC cells were treated with different concentrations of indicated compounds for 72 h. Cell viability was evaluated by MTS assay. See Supplementary Table 1 for the structure of the compounds. b Chemical structures of RYL-687. c Seven lung cancer cell lines were treated with different concentrations of RYL-687 for 48 h, and cell viability was evaluated by MTS assay and is shown as relative viability compared with the control. d H460 and A549 cells were treated with the indicated concentrations of RYL-687 for the indicated time points, and cell viability was determined by MTS assay.
RYL-687 significantly inhibits lung cancer cells with wt p53
H460 and A549 cells bear wt p53, whereas H1975 and H1299 lines harbor R273H and deletion mutations of p53, respectively. The GI50 values for H460 and A549 cells were much lower than that for H1975 and H1229 cells (Supplementary Table 1), suggesting a potential role for p53 to play in anti-tumor effects of RYL-687. To confirm this possibility, three cell lines with p53 mutations, i.e., H520 with W146*, HCC827 with V218del, and HOP-62 with splice site mutation, were also treated with RYL-687. We found that among the 7 NSCLC lines upon 10–5000 nM RYL-687 treatment for 48 h, A549 and H460 cells were more sensitive to this compound than the other 5 cell lines (Fig. 1c). RYL-687 inhibited the growth of H460 and A549 cells in a time- and dose-dependent manner (Fig. 1d). Based on the above data, RYL-687 was chosen as the representative molecule for the following experiments to investigate its anticancer activity and its effects on p53 expression.
RYL-687 potently induces the expression of p53 in NSCLC cells with wt p53
We investigated the effects of RYL-687 on p53. Without drug treatment, the expression of p53 was undetectable in H460, A549 and H1299 cells, and was relatively high in H1975 cells (Fig. 2a). Interestingly, RYL-687 drastically induced, in a dose-dependent manner, upregulation of p53 in A549 and H460 but not H1975 and H1299 cells (Fig. 2a). As a transcription factor, p53 exerted its functions by modulating the transcription of target genes in the nuclei [23]. We determined the nuclear/cytoplasmic distribution of p53 by Western blot, and reported that RYL-687 promoted p53 nuclear accumulation in H460 cells (Fig. 2b). Consistently, immunofluorescence analysis suggested that p53 mainly accumulated in the nucleus of the cells upon RYL-687 treatment, in both time- and dose-dependent manners (Fig. 2c). Nutlin3A [24] and cisplatin (DDP) [25] are two typical activators of p53. The effect of RYL-687 on p53 was compared with that of Nutlin3A and DDP in H460 and A549 cells, and the results showed that treatment with RYL-687 at 10 nM for 12–24 h drastically upregulated p53, at an extent equal to or even stronger than that induced by Nutlin3A or DDP at 10 μM for 12–24 h (Fig. 2d, e). These observations indicated that the positive effect of RYL-687 on p53 may be stronger than that of Nutlin3A and DDP.
a A549, H460, H1975 and H1299 cells were treated with the indicated concentrations of RYL-687 for 6 h, harvested, and subjected to Western blot analysis using the indicated antibodies. b After H460 cells were treated with RYL-687 for 6 h, nuclear and cytoplasmic protein fractions were prepared and evaluated by Western blotting. c Immunofluorescence assays of H460 cells using antibodies against p53 (green) and 4’,6-diamidino-2-phenylindole (DAPI) to counterstain the nucleus (blue) after treatment with RYL-687 at the indicated concentrations for the indicated time points. H460 (d) and A549 (e) cells were treated with RYL-687 (10 nM), Nutlin3A (10 μM), and cisplatin (DDP) (10 μM) at the indicated time points and lysed for Western blot analysis. f H460 cells were pre-treated with RYL-687 at 10 nM for 6 h to increase p53 to a detectable level, and then further treated with CHX (50 μg/mL) in the absence or presence of RYL-687 (10 nM), and the expression of p53 expression was evaluated by Western blotting. Numbers under the Western blot bands are the relative expression values to Actin determined by densitometry analysis. g H1299 cells were transfected with wt p53, treated with RYL-687, and analysed for cell viability. h H1975 cells were transfected with CRISPR-Cas9 system to silence the mutant p53, puromycin resistance cells were selected and transfected with wt p53, followed by treatment with RYL-687 and analysis of cell viability. **P < 0.01; ***P < 0.001; ****P < 0.0001.
We tested the effects of RYL-687 on p53 stability using the protein synthesis inhibitor cycloheximide (CHX). In the absence and presence of CHX (50 µg/mL), p53 was undetectable in H460 cells (Fig. 2f). Therefore, H460 cells were pre-treated with RYL-687 at 10 nM for 6 h to increase p53 to a detectable level, and then further treated with CHX in the absence or presence of RYL-687. We reported that without RYL-687 treatment, the upregulated p53 was downregulated by CHX treatment; however, in the presence of RYL-687, CHX-induced p53 downregulation was markedly inhibited (Fig. 2f). These results demonstrated that RYL-687 causes the accumulation of p53 by enhancing its protein stability.
p53 is critical to the anti-lung cancer activity of RYL-687
To evaluate the role of wt p53 in RYL-687-induced inhibitory effects on NSCLC cells, TP53 was transfected into H1299 cells which were then treated with the compound. We showed that while H1299 cells were relatively resistant to the compound, ectopic expression of wt TP53 significantly enhanced inhibition of H1299 cell viability caused by RYL-687 at 0.1–20 nM (Fig. 2g). In H1975 cells, knockdown of mutant TP53 by CRISPR-Cas9 system potentiated RYL-687-induced suppression of cell viability, and overexpression of wt TP53 further enhanced this effect (Fig. 2h). These results demonstrated that the wt p53 is critical to the anti-lung cancer activity of RYL-687.
Effects of RYL-687 on p53 target genes and cell fate
p53 regulates the expression of hundreds of genes that are involved in various biological processes, including in DNA damage repair, cell cycle arrest, apoptosis and senescence [26]. p21 is an important downstream targets of p53 [27]. We showed that in H460 and A549 cells, RYL-687 upregulated p21 in a dose-dependent manner (Fig. 3a). Upon RYL-687 treatment, p53 targets Bax and MDM2 [28] were upregulated at protein and mRNA levels (Fig. 3a, b). RYL-687 slightly upregulated TP53 at mRNA level (Fig. 3b). RYL-687 downregulated CDK2, CDK4, Cyclin A2 (Fig. 3c), and arrested cell cycle at S phase (Fig. 3d). On the other hand, RYL-687 downregulated Bcl-2 and upregulated Bax (Fig. 3e, upper panel). RYL-687 increased cytosolic Cytochrome c (Cyt c) with decreased mitochondrial Cyt c (Fig. 3e, lower panel), and activated Casp-9 and Casp-3 with cleavage of poly (ADP-ribose) polymerase (PARP) (Fig. 3e, upper panel), leading to programmed cell death (Fig. 3f). RYL-687 also significantly inhibited colony forming activity of the cells (Fig. 3g). These data demonstrated that RYL-687 had selective cytotoxicity to lung cancer cells and can induce S-phase arrest and cell apoptosis in NSCLC cells with wt p53.
a The cells were treated with RYL-687 at the indicated concentrations for 12 h, lysed, and subjected to immunoblotting using the indicated antibodies. b The mRNA levels of TP53, MDM2 and Bax were analyzed by quantitative RT-PCR, in H460 and A549 cells that were treated with RYL-687 at 10 nM for 12 h. c, d H460 and A549 cells were treated with the indicated concentrations of RYL-687 for 24 h. The cells were lysed for Western blot analysis using the indicated antibodies (c), or analysed by flow cytometry to evaluate the cell cycle distribution (d). e, f The cells were treated with the indicated concentrations of RYL-687 for 24 h, and lysed for Western blot analysis using total or cytosolic/mitochondrial proteins (e), or analysed by flow cytometry analysis using Annexin V-FITC/PI staining for apoptotic cell death (f). g Colony formation assays were performed in H460 and A549 cells with the indicated concentrations of RYL-687. Representative images are shown. *P < 0.05, **P < 0.01; ***P < 0.001 and ****P < 0.0001.
Identification of NQO1 as a RYL-687-binding protein
To reveal the molecular mechanism of the RYL-687-induced upregulation of p53 in NSCLC cells, Bio-687 (Supplementary Fig. 1a–c) was synthesized and used to perform a pull-down assay. In contrast to the biotin-treated samples, a specific protein band was observed in the protein samples of Bio-687-treated cells that were separated by polyacrylamide gels and detected by silver-staining (Fig. 4a). Mass spectrometry analysis of the protein bands showed that the NQO1 was one of the Bio-687-binding proteins and was identified each time in the triplicate experiments (Table 1). NQO1, a cytosolic oxidoreductase that protected cells against oxidative stress and can stabilize p53 by inhibiting its 20S proteasomal degradation [29], was chosen to investigate the mechanism of action of RYL-687-induced p53 expression in NSCLC cells.
a Silver staining of proteins precipitated with Biotin and Bio-687 at 10 μM for 6 h in H460 cells. b H460 cells were transfected with or without siNQO1, treated with Biotin or Bio-687 at 10 μM for 6 h and lysed, and the cell lysates were subjected to immunoprecipitation using streptavidin agarose and Western blotting using indicated antibodies. c H460 cells were treated with RYL-687 at 10 nM for 6 h, lysed, and subjected to immunoprecipitation and immunoblotting using indicated antibodies. d The expression of p53 and NQO1 in H460 and A549 cells treated with vehicle control (DMSO) or RYL-687 at 10–50 nM for 12 h. The cells were harvested and detected by immunofluorescence assays with indicated antibodies. e H460 cells were treated with RYL-687 at 10 nM for 6–12 h and lysed, RNA was extracted, and NQO1 expression was evaluated by quantitative RT-PCR. P values, Student’s t test, *P < 0.05; **P < 0.01, ***P < 0.001. f Western blot analysis of NQO1, Nrf2 and p53 expression in H460 and A549 cells treated with RYL-687 for 12 h. g H460 and A549 cells were incubated with RYL-687 for 24 h. ROS detection was performed with DCFH-DA dye in different groups by flow cytometry. h H460 and A549 cells were treated with the indicated concentrations of RYL-687 for 12 h, lysed, and analysed by Western blot using indicated antibodies.
RYL-687 promotes NQO1-p53 interaction and reactive oxygen species production
The interaction between Bio-687 and NQO1 was verified by co-immunoprecipitation and Western blot analysis using streptavidin-agarose and an anti-NQO1 antibody (Fig. 4b). p53 but not β-Actin was also pulled down by streptavidin agarose (Fig. 4b, left panel). To determine the Bio-687-p53 interaction represents a direct or indirect (through NQO1) binding, H460 cells were transfected with siNQO1 and treated with Bio-687. We showed that p53 was pulled down by Bio-687, while silencing of NQO1 abrogated p53-Bio-687 interaction (Fig. 4b, right panel), indicating that Bio-687 indirectly binds p53 through NQO1. In cells treated with RYL-687, NQO1 could be pulled down by anti-p53 monoclonal antibody (Fig. 4c), suggesting the interaction between p53 and NQO1. Immunofluorescence analysis showed that RYL-687 increased the expression levels of both the NQO1 and p53 in H460 and A549 cells in a dose-dependent manner (Fig. 4d). Moreover, co-localization of NQO1 and p53 in the nucleus was observed within the cells upon RYL-687 treatment (Fig. 4d). RYL-687-induced upregulation of NQO1 was seen at both the mRNA (Fig. 4e) and protein (Fig. 4f) levels in H460 and A549 cells.
It has been demonstrated that activation of NQO1 can cause DNA damage and cell death, which is associated with accumulation of reactive oxygen species (ROS) in the cells. The increase of NQO1 can further promote the production of ROS [30]. Therefore, the intracellular ROS level was detected using a 2,7-dichloro-fluorescin diacetate (DCFH-DA) assay. We showed that RYL-687 increased intracellular ROS levels in H460 and A549 cells (Fig. 4g). Moreover, the protein level of Nrf2 was upregulated by RYL-687 treatment (Fig. 4f). Studies showed that elevated γ-H2AX expression was associated with ROS-mediated DNA damage [31]. We reported that the expression of γ-H2AX in H460 and A549 cells was also upregulated by RYL-687 in a dose-dependent manner (Fig. 4h).
NQO1 is required for RYL-687-induced upregulation of p53
Uridine is a compound that can inhibit the synthesis of pyrimidine as well as the expression of NQO1 [32]. To elucidate the role of NQO1 on RYL-687-induced suppression of NSCLC cells and p53 upregulation, uridine was used as an inhibitor of NQO1. As shown in Fig. 5a, the inhibitory effect of RYL-687 on H460 cell proliferation was significantly suppressed by uridine (50 μM). Moreover, uridine also rescued the S-phase arrest induced by RYL-687 in both H460 and A549 cells (Fig. 5b). Knockdown of NQO1 by siRNA led to a significant inhibition of p53 upregulation induced by RYL-687 (Fig. 5c). Furthermore, co-treatment of H460 and A549 cells with uridine for 24 h drastically repressed the upregulation of NQO1, p53, MDM2 and p21 proteins by RYL-687 (Fig. 5d). These results indicated that NQO1 was required for RYL-687-induced upregulation of p53.
a H460 cells were exposed to RYL-687 and RYL-687 in combination with uridine (50 μM) for 48 h. Cell viability was evaluated by MTS assay. b H460 and A549 cells were treated with RYL-687 and RYL-687 plus uridine for 24 h. The cells were analysed by flow cytometry to evaluate the cell cycle distribution. c H460 cells were transfected with siNQO1, treated with RYL-687 at 10 nM for 12 h, and lysed for Western blot analysis using indicated antibodies. d Western blot analysis of p53 levels in H460 and A549 cells treated with RYL-687 and/or uridine (50 μM) for 12 h.
The Prodrug exhibits potent anti-lung cancer activity in vivo
The water solubility of RYL-687 was poor and to investigate the anti-lung cancer activity of RYL-687, the RYL-687-related prodrug (Supplementary Table 1) was synthesized. In NOD-SCID mice inoculated with 1 × 106 H460 cells, treatment with prodrug at 20 or 60 mg/kg (once per day for 3 weeks) significantly inhibited tumor growth and tumor weight (Fig. 6a–c). Methotrexate (MTX), one of the most widely used anticancer agents [33], also significantly inhibited tumor growth (Fig. 6a–c). Prodrug did not reduce the body weight of the mice (Fig. 6d). We further tested the efficacy of the prodrug on an NSCLC patient-derived xenograft (PDX) mouse model, in which the patient sample harbored wt p53 as revealed by whole exome sequencing. We found that treatment with Prodrug at 20–60 mg·kg−1·d−1 for 20 days significantly inhibited tumor growth (Fig. 6e–g) but did not reduce the body weight of the mice (Fig. 6h).
a H460 (1 × 106) cells were inoculated subcutaneously into the right flank of the NOD-SCID mice, which were treated with indicated concentrations of prodrug and MTX. Tumor volume was estimated every 2 days. Data are shown as mean ± SD. b Images of xenograft tumors isolated from the mice. c Weights of xenograft tumors isolated from the mice. d Body weights of the mice. e Patient-derived xenograft mouse model was established using NOD-SCID mice and a wt-p53-bearing NSCLC tumor sample and treated with prodrug and MTX, and tumor volume was estimated every 2 days. Data are shown as mean ± SD. n = 5 for each group. f Images of xenograft tumors obtained from the mice. g Weights of xenograft tumors isolated from the mice. h Body weights of the mice. CDX cell-derived xenograft, PDX patient-derived xenograft. *P < 0.05; **P < 0.01.
Discussion
The tumor suppressor p53 plays an important role in protecting the genome and preventing cell transformation [34]. Generally, cancer cells with wt p53 can reactivate downstream pathways that mediate cell death [35]. Therefore, restoring the activity of wt p53 may be a very promising strategy for cancer treatment. RYL-687 is a new quinolone compound derived from RYL-634 that has been shown to have broad-spectrum antiviral activities [18]. Here we show that RYL-687 exhibits potent inhibitory effects on NSCLC cells with GI50 values of around 23 nM on NSCLC cells with wt p53. RYL-687 induces production of ROS and upregulation of Nrf2 and NQO1, leading to upregulation of p53 and its downstream targets including p21 and Bax. As a consequence, cell cycle is arrested at S phase and apoptosis is induced in NSCLC cells (Fig. 7). Therefore, RYL-687 is a potent reactivator of wt p53.
NQO1 is a cytosolic flavoenzyme that catalyses the obligatory two-electron reduction of quinone substrates, using both NADH and NADPH as electron donors. Upon bioreductive activation of certain quinones, the corresponding unstable hydroquinones rapidly react with physiological oxygen in cells to provide 2 equivalents of superoxide, which is the main constituent of ROS, and regenerate the quinones [36]. These redox substrates rapidly catalyse and produce large amounts of toxic ROS in cells overexpressing NQO1 to kill cancer cells [37]. NQO1 also has non-enzymatic functions in stabilizing a number of cellular regulators including p53 [32]. The expression of NQO1 is low in normal tissues; but in cancer the expression and activity of NQO1 is complicated. NQO1 C609T (P187S) polymorphism, which results in reduced NQO1 activity, is associated with a predisposition to cancers (in particular, bladder and gastric cancers) [38], and this link may be attributed to its less effectiveness in stabilizing p53 [39]. The expression of NQO1 is significantly upregulated in colon, pancreatic and breast cancers [40, 41]. In NSCLC, NQO1 activity is drastically higher in tumors than in normal lung samples [42]. The large differential expression/activity of NQO1 between tumors and counterpart normal tissues suggests that NQO1 could be a potential target for cancer therapy [43], and some NQO1 substrates have been reported to be potential antitumor drugs [37]. We reported that RYL-687 induced a positive feedback loop to facilitate NQO1 function. On one hand, RYL-687 induced production of ROS and DNA damage to activate Nrf2, which upregulated NQO1 to stabilize p53. On the other hand, NQO1 enhances generation of ROS, which further increased Nrf2 to potentiate the effect of RYL-687. In addition, RYL-687 binds to NQO1, facilitating the formation of NQO1-p53 complex (Fig. 7). These data demonstrate that RYL-687 represents a novel NQO1 modulator. Natural compound curcumin can also target NQO1 to stabilize p53, at a concentration (20,000 nM) that is much higher than RYL-687 [44]. Thus, targeting NQO1 could be a rational strategy to restore wt p53 for cancer treatment.
p53 is usually kept inactive by the ubiquitination and subsequent degradation in the 26S proteasome mediated by a number of ubiquitin E3 ligases such as MDM2, Pirh2, COP1, and others [45,46,47,48,49]. This kind of degradation is ubiquitin-dependent (UD) [29], whereas an ubiquitin-independent (UI) degradation approach is also uncovered in the 20S proteasome [50,51,52]. The UD and UI modes of degradation that are of equal importance with distinct kinetics, enable a differential accumulation pattern of p53 following different stressors [52, 53]. For the UI mode of degradation, NQO1 binds the N terminus of p53 and protects it from the 20S proteasomal degradation [54]. However, the “hot-spot” mutations of p53, e.g., R175H, R248H, and R273H, are resistant to NQO1 inhibitor dicoumarol-induced degradation, and arginines at positions 175 and 248 are essential for dicoumarol-induced p53 degradation [55]. A double negative feedback loop between NQO1 and the 20S proteasome has been reported, whereby NQO1 prevents the proteolytic activity of the 20S proteasome and the 20S proteasome degrades the apo form of NQO1 [52]. Thus, NQO1 has anti-tumor activity, regardless of its enzymatic activity [56]. In our study, we found that NSCLC cells with wt p53 were more sensitive to RYL-687 than those with mutant p53. Inhibition of NQO1 by siNQO1 and NQO1 inhibitor uridine significantly suppressed RYL-687-induced p53 upregulation and its inhibitory effects on the NSCLC cells with wt p53, indicating that NQO1 was essential for RYL-687’s anti-NSCLC activity. These results also provided explanation for the findings that NSCLC cells with mutant p53 were insensitive to RYL-687, because the mutant p53 were resistant to the 20S proteasomal degradation.
In summary, our results demonstrated that the novel quinolone compound RYL-687 exerts potent anti-NSCLC activity in vitro and in vivo, through induction of cell cycle arrest at S phase, promotion of apoptosis, and inhibition of colony forming activity of NSCLC cells with wt p53. This compound upregulated the expression of p53 in cells with wt but not mutant p53, via induction of NQO1 expression that is able to inhibit wt p53 degradation by 20S proteasome in a UI pathway. p53 is maintained wt in about a half of the cancer patients, suggesting that reactivation of wt p53 could be a promising therapeutic strategy in fighting this deadly disease. Therefore, more studies should be carried out to investigate this NQO1-p53 targeting approach and optimize the property of RYL-687 for possible clinical application.
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33.
Howlader N, Forjaz G, Mooradian MJ, Meza R, Kong CY, Cronin KA, et al. The effect of advances in lung-cancer treatment on population mortality. N Engl J Med. 2020;383:640–9.
Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–54.
Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr., Wu YL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34.
Zhou G. Tobacco, air pollution, environmental carcinogenesis, and thoughts on conquering strategies of lung cancer. Cancer Biol Med. 2019;16:700–13.
You QD, Li ZY, Huang CH, Yang Q, Wang XJ, Guo QL, et al. Discovery of a novel series of quinolone and naphthyridine derivatives as potential topoisomerase I inhibitors by scaffold modification. J Med Chem. 2009;52:5649–61.
Foti JJ, Devadoss B, Winkler JA, Collins JJ, Walker GC. Oxidation of the guanine nucleotide pool underlies cell death by bactericidal antibiotics. Science. 2012;336:315–9.
Dopp E, Yadav S, Ansari FA, Bhattacharya K, Von Recklinghausen U, Rauen U, et al. ROS-mediated genotoxicity of asbestos-cement in mammalian lung cells in vitro. Part Fibre Toxicol. 2005;2:9.
Halliwell B, Cross CE. Oxygen-derived species: their relation to human disease and environmental stress. Environ Health Perspect. 1994;102:5–12.
Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–50.
Herold C, Ocker M, Ganslmayer M, Gerauer H, Hahn EG, Schuppan D. Ciprofloxacin induces apoptosis and inhibits proliferation of human colorectal carcinoma cells. Br J Cancer. 2002;86:443–8.
Seo K, Holt R, Jung YS, Rodriguez CO Jr., Chen X, Rebhun RB. Fluoroquinolone-mediated inhibition of cell growth, S-G2/M cell cycle arrest, and apoptosis in canine osteosarcoma cell lines. PLoS One. 2012;7:e42960.
Paul M, Gafter-Gvili A, Fraser A, Leibovici L. The anti-cancer effects of quinolone antibiotics? Eur J Clin Microbiol Infect Dis. 2007;26:825–31.
Lin MS, Hong TM, Chou TH, Yang SC, Chung WC, Weng CW, et al. 4(1H)-quinolone derivatives overcome acquired resistance to anti-microtubule agents by targeting the colchicine site of beta-tubulin. Eur J Med Chem. 2019;181:111584.
Yadav V, Talwar P. Repositioning of fluoroquinolones from antibiotic to anti-cancer agents: an underestimated truth. Biomed Pharmacother. 2019;111:934–46.
Bykowska A, Komarnicka UK, Jezowska-Bojczuk M, Kyziol A. Cu(I) and Cu(II) complexes with phosphine derivatives of fluoroquinolone antibiotics - a comparative study on the cytotoxic mode of action. J Inorg Biochem. 2018;181:1–10.
Yang Y, Cao L, Gao H, Wu Y, Wang Y, Fang F, et al. Discovery, optimization, and target identification of novel potent broad-spectrum antiviral inhibitors. J Med Chem. 2019;62:4056–73.
Zhang K, Chen D, Ma K, Wu X, Hao H, Jiang S. NAD(P)H:quinone oxidoreductase 1 (NQO1) as a therapeutic and diagnostic target in cancer. J Med Chem. 2018;61:6983–7003.
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308.
Liu Z, Ma L, Wen ZS, Hu Z, Wu FQ, Li W, et al. Cancerous inhibitor of PP2A (CIP2A) is targeted by natural compound celastrol for degradation in non-small-cell lung cancer. Carcinogenesis. 2014;35:905–14.
Reis AC, Kolvenbach BA, Nunes OC, Corvini PFX. Biodegradation of antibiotics: the new resistance determinants - part II. N Biotechnol. 2020;54:13–27.
Khoo KH, Verma CS, Lane DP. Drugging the p53 pathway: understanding the route to clinical efficacy. Nat Rev Drug Discov. 2014;13:217–36.
Dong G, He S, Qin X, Liu T, Jiang Y, Li X, et al. Discovery of nonpeptide, environmentally sensitive fluorescent probes for imaging p53-MDM2 interactions in living cell lines and tissue slice. Anal Chem. 2020;92:2642–8.
Jing L, Song F, Liu Z, Li J, Wu B, Fu Z, et al. MLKL-PITPalpha signaling-mediated necroptosis contributes to cisplatin-triggered cell death in lung cancer A549 cells. Cancer Lett. 2018;414:136–46.
Hafner A, Bulyk ML, Jambhekar A, Lahav G. The multiple mechanisms that regulate p53 activity and cell fate. Nat Rev Mol Cell Biol. 2019;20:199–210.
Eskandari M, Shi Y, Liu J, Albanese J, Goel S, Verma A, et al. The expression of MDM2, MDM4, p53 and p21 in myeloid neoplasms and the effect of MDM2/MDM4 dual inhibitor. Leuk Lymphoma. 2021;62:167–75.
Chen S, Wu JL, Liang Y, Tang YG, Song HX, Wu LL, et al. Arsenic trioxide rescues structural p53 mutations through a cryptic allosteric site. Cancer Cell. 2021;39:225–39.e8.
Asher G, Tsvetkov P, Kahana C, Shaul Y. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 2005;19:316–21.
Al-Gubory KH, Fowler PA, Garrel C. The roles of cellular reactive oxygen species, oxidative stress and antioxidants in pregnancy outcomes. Int J Biochem Cell Biol. 2010;42:1634–50.
Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–68.
Khutornenko AA, Roudko VV, Chernyak BV, Vartapetian AB, Chumakov PM, Evstafieva AG. Pyrimidine biosynthesis links mitochondrial respiration to the p53 pathway. Proc Natl Acad Sci USA. 2010;107:12828–33.
Widemann BC, Adamson PC. Understanding and managing methotrexate nephrotoxicity. Oncologist. 2006;11:694–703.
Lavin MF, Gueven N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006;13:941–50.
Goodwin EC, Dimaio D. Repression of human papillomavirus oncogenes in HeLa cervical carcinoma cells causes the orderly reactivation of dormant tumor suppressor pathways. Proc Natl Acad Sci USA. 2000;97:12513–8.
Newsome JJ, Colucci MA, Hassani M, Beall HD, Moody CJ. Benzimidazole- and benzothiazole-quinones: excellent substrates for NAD(P)H:quinone oxidoreductase 1. Org Biomol Chem. 2007;5:3665–73.
Wu X, Li X, Li Z, Yu Y, You Q, Zhang X. Discovery of nonquinone substrates for NAD(P)H: quinone oxidoreductase 1 (NQO1) as effective intracellular ROS generators for the treatment of drug-resistant non-small-cell lung cancer. J Med Chem. 2018;61:11280–97.
Lajin B, Alachkar A. The NQO1 polymorphism C609T (Pro187Ser) and cancer susceptibility: a comprehensive meta-analysis. Br J Cancer. 2013;109:1325–37.
Pey AL, Megarity CF, Timson DJ. NAD(P)H quinone oxidoreductase (NQO1): an enzyme which needs just enough mobility, in just the right places. Biosci Rep. 2019;39:BSR20180459.
Ma Y, Kong J, Yan G, Ren X, Jin D, Jin T, et al. NQO1 overexpression is associated with poor prognosis in squamous cell carcinoma of the uterine cervix. BMC Cancer. 2014;14:414.
Yang Y, Zhang Y, Wu Q, Cui X, Lin Z, Liu S, et al. Clinical implications of high NQO1 expression in breast cancers. J Exp Clin Cancer Res. 2014;33:14.
Malkinson AM, Siegel D, Forrest GL, Gazdar AF, Oie HK, Chan DC, et al. Elevated DT-diaphorase activity and messenger RNA content in human non-small cell lung carcinoma: relationship to the response of lung tumor xenografts to mitomycin Cl. Cancer Res. 1992;52:4752–7.
Park MT, Song MJ, Lee H, Oh ET, Choi BH, Jeong SY, et al. beta-lapachone significantly increases the effect of ionizing radiation to cause mitochondrial apoptosis via JNK activation in cancer cells. PLoS One. 2011;6:e25976.
Patino-Morales CC, Soto-Reyes E, Arechaga-Ocampo E, Ortiz-Sanchez E, Antonio-Vejar V, Pedraza-Chaverri J, et al. Curcumin stabilizes p53 by interaction with NAD(P)H:quinone oxidoreductase 1 in tumor-derived cell lines. Redox Biol. 2020;28:101320.
Chao CC. Mechanisms of p53 degradation. Clin Chim Acta. 2015;438:139–47.
Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303.
Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–9.
Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–91.
Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92.
Asher G, Lotem J, Sachs L, Kahana C, Shaul Y. Mdm-2 and ubiquitin-independent p53 proteasomal degradation regulated by NQO1. Proc Natl Acad Sci USA. 2002;99:13125–30.
Camus S, Menendez S, Cheok CF, Stevenson LF, Lain S, Lane DP. Ubiquitin-independent degradation of p53 mediated by high-risk human papillomavirus protein E6. Oncogene. 2007;26:4059–70.
Moscovitz O, Tsvetkov P, Hazan N, Michaelevski I, Keisar H, Ben-Nissan G, et al. A mutually inhibitory feedback loop between the 20S proteasome and its regulator, NQO1. Mol Cell. 2012;47:76–86.
Tsvetkov P, Reuven N, Prives C, Shaul Y. Susceptibility of p53 unstructured N terminus to 20 S proteasomal degradation programs the stress response. J Biol Chem. 2009;284:26234–42.
Dinkova-Kostova AT, Talalay P. NAD(P)H:quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch Biochem Biophys. 2010;501:116–23.
Asher G, Lotem J, Tsvetkov P, Reiss V, Sachs L, Shaul Y. P53 hot-spot mutants are resistant to ubiquitin-independent degradation by increased binding to NAD(P)H:quinone oxidoreductase 1. Proc Natl Acad Sci USA. 2003;100:15065–70.
Oh ET, Park HJ. Implications of NQO1 in cancer therapy. BMB Rep. 2015;48:609–17.
Acknowledgements
This work was jointly supported by the National Key Research and Development Program of China (Nos. 2020YFA0803300), the CAMS Innovation Fund for Medical Sciences (CIFMS; No. 2019-I2M-1–003), the Key Project of the National Natural Science Foundation of China (No. 81830093), the National Natural Science Funds for Distinguished Young Scholar (No. 81425025), and the National Natural Science Foundation of China (Nos. 81672765 and 81802796).
Author information
Authors and Affiliations
Contributions
The project was conceived by GBZ and YR. The experiments were designed by GBZ, YR. The experiments were conducted by HY, HYG, HG, GZW, YQY, QH, LJL, QZ, and DWX. Data were analyzed by YR, GBZ. The manuscript was written by HY, and GBZ.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Yu, H., Gao, Hy., Guo, H. et al. Upregulation of wild-type p53 by small molecule-induced elevation of NQO1 in non-small cell lung cancer cells. Acta Pharmacol Sin 43, 692–702 (2022). https://doi.org/10.1038/s41401-021-00691-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-021-00691-8