Abstract
Guangsangon E (GSE) is a novel Diels–Alder adduct isolated from leaves of Morus alba L, a traditional Chinese medicine widely applied in respiratory diseases. It is reported that GSE has cytotoxic effect on cancer cells. In our research, we investigated its anticancer effect on respiratory cancer and revealed that GSE induces autophagy and apoptosis in lung and nasopharyngeal cancer cells. We first observed that GSE inhibits cell proliferation and induces apoptosis in A549 and CNE1 cells. Meanwhile, the upregulation of autophagosome marker LC3 and increased formation of GFP–LC3 puncta demonstrates the induction of autophagy in GSE-treated cells. Moreover, GSE increases the autophagy flux by enhancing lysosomal activity and the fusion of autophagosomes and lysosomes. Next, we investigated that endoplasmic reticulum (ER) stress is involved in autophagy induction by GSE. GSE activates the ER stress through reactive oxygen species (ROS) accumulation, which can be blocked by ROS scavenger NAC. Finally, inhibition of autophagy attenuates GSE-caused cell death, termed as “autophagy-mediated cell death.” Taken together, we revealed the molecular mechanism of GSE against respiratory cancer, which demonstrates great potential of GSE in the treatment of representative cancer.
Similar content being viewed by others
Introduction
Lung cancer and nasopharyngeal carcinoma (NPC), representative respiratory cancers, are the most frequently diagnosed cancers and the leading cause of cancer death worldwide and especially in China [1]. Non-small cell lung cancer (NSCLC), which represents the remaining 85% of lung cancers, has a lower 5-year overall survival rate of 15% [2, 3]. A review of the outcome of 57,496 international screening cases substantiated the indolent behavior of nonsolid pulmonary nodules [4], suggesting that a more conservative approach to surgical resection is appropriate [5]. However, the overall cure and survival rates of NSCLC remain low, particularly in metastatic disease [6]. NPC is the most common malignant tumor of the head and neck. According to the International Agency for Research on Cancer, there were ~129,000 new cases of NPC in 2018, accounting for 0.7% of all cancers, and more than 70% of these cases occurred in East and Southeast Asia [7]. NPC patients also face the same low cure and survival rates. Thus, the need to develop novel therapeutics to expand clinical benefits to more patients with respiratory cancer and improve their outcomes is urgent.
Morus alba L. contains various bioactive compounds, such as polyphenols and anthocyanins, and is commonly used as a medicinal herb. In ancient China, the leaves of Morus alba L. were often prescribed to relieve cough. Previous phytochemical investigations showed that Morus alba L. contains a large number of biologically important secondary metabolites, including 2-arylbenzofurans, stilbenes, flavonoids, and Diels–Alder adducts [8, 9]. These compounds exhibit various biological properties, such as cytotoxic, antioxidant, anti-inflammatory, antibacterial, antimicrobial, and antiviral properties [10]. More studies have shown that Morus alba L. extracts have anticancer activity and that the intake of these extracts inhibits cancer growth [11]. For example, in lung cancer, the root bark of Morus alba L. inhibits the migratory activity of cancer cells [12]; in prostate cancer, morusin induces cytotoxicity and autophagy in cancer cells, and the inhibition of autophagy affects the cytotoxicity of morusin [13]. In addition, autophagy induction is involved in the anti-inflammatory effect of moracin M, which inhibits LPS-induced PI3K-AKT activation through autophagy [14], and the antioxidant activity of sanggenon C, which protects against cardiomyocyte hypoxia injury by autophagy [15]. Here, we isolated and purified another effective component of the leaves of Morus alba L., guangsangon E (GSE). Its structure is shown in Fig. 1a. To date, there have been no reports on the biological activity and molecular mechanism of GSE. In the present study, we aimed to investigate the anticancer effect and molecular mechanism of GSE in respiratory cancer.
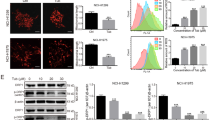
a The structure of GSE. b Cell viability of A549 and CNE1 cells under GSE treatment for different times. c Colony formation ability of A549 and CNE1 cells after GSE treatment. d as in c, Colony numbers were determined and statistically analyzed.**P < 0.01.e A549 cells were treated with GSE as indicated, and cell apoptosis was measured using Annexin V/PI staining coupled with flow cytometry. **P < 0.01. f The expression of cleaved caspase 3 in GSE-treated cells was determined using Western blotting. g Analysis of Bax and Bcl-2 expression in A549 cells treated with GSE was carried out using immunofluorescence. h The ratio of Bcl-2 to Bax was calculated and statistically analyzed. **P < 0.01.
Autophagy is a ubiquitous catabolic process that involves the degradation of cytoplasmic components, including proteins, cytoplasm and even whole organelles, via the lysosomal pathway [16, 17]. Autophagy has historically been considered a defensive mechanism in response to various stresses because it is responsible for metabolic compensation and organelle turnover [18]. Generally, autophagy induction refers to an increase in autophagic flux (completion of autophagy) rather than the presence of increased autophagy markers in dying cells [16, 19]. The role of autophagy in tumorigenesis is very complicated. Multiple studies have demonstrated that autophagy participates in anticancer drug resistance mechanisms and protects lung cancer cells from therapeutics [20,21,22,23]. In contrast, a study [24] suggested that autophagy acts to sensitize lung cancer cells to therapeutic drugs. It has been proposed that autophagy functions as a cytotoxic mechanism when cell death mediated through apoptosis is compromised [25]. Thus, it is necessary to reveal the biological function of autophagy in lung cancer, which will be helpful to develop novel therapeutics targeting autophagy.
In this study, we found that GSE induced autophagy and apoptosis in A549 and CNE1 cells. GSE treatment caused the unfolded protein response (UPR) through oxidative stress, which further led to autophagy. Interestingly, autophagy and apoptosis are two parallel pathways of GSE-induced cell death. These findings indicate that GSE might be a novel therapeutic agent for respiratory cancers.
Materials and methods
Reagents and antibodies
The following reagents were used in our research: N-acetyl-cysteine (NAC, Aladdin, #616-91-1), crystal violet (Beyotime, #C0121), MTT (Solarbio, #M8180), dimethyl sulfoxide (DMSO, SCR, #30072418), a BCA protein assay kit (Solarbio, #PC0020), si-Atg5 (CST, #6345), si-IRE1α (GenePharma, Shanghai, 5′-CUACUGGAUGAUAAAUUUGCUUCATT-3′), negative control (GenePharma, 5′-UUCUCCGAACGUGUCACGUTT-3′), chloroquine (CQ, Sigma-Aldrich, #C6628), LysoTracker Red DND-99 (Invitrogen, #L7528), endoplasmic reticulum (ER) Tracker green (CST, #8787S), ER Tracker red (Beyotime, #C1041), Fluo-3 AM (Beyotime, #S1056), fetal bovine serum (FBS, Biological Industries, #04-001-1A), trypsin with EDTA (Genom, Hangzhou, #GNM-25200), trypsin (Genom, Hangzhou, #GNM-27250), a reactive oxygen species (ROS) assay kit (NJJCTECH, #E004-1-1), an Annexin V-FITC/PI apoptosis kit (KeyGenBiotech, #KGA105), DAPI (Solarbio, #C0065), BeyoECL Plus (Beyotime, #P0018S), a mitochondrial membrane potential (MMP) assay kit with JC-1 (NJJCTECH, #G009-1-2), and Lipofectamine 2000 reagent (Invitrogen, #11668027).
All of the human antibodies against the following used in the experiments were from Cell Signaling Technology (CST): light chain 3 (LC3) (CST, #3868), caspase 3 (CST, #14220), cleaved caspase 3 (CST, #9661), GAPDH (CST, #5174), ATG5 (CST, #12994), inositol-requiring enzyme 1α (IRE1α) (CST, #3294), glucose-regulated protein 78 (GRP78) (CST, #3177), activating transcription factor 4 (ATF4) (CST, #11815), Bcl-2 (CST, #4223), and Bax (CST, #5023). The secondary antibodies goat anti-rabbit and goat anti-mouse IgG-HRP were obtained from Immuno Research Laboratories (#111-035-003 and #115-035-003, respectively).
GSE with a high chemical purity of more than 98% was isolated and purified by Professor Jing-kui Tian from the Key Laboratory of Biomedical Engineering at Zhejiang University. Its content in mulberry leaves had been further improved through UV induction technology.
Cell culture
The A549 human non-small-cell lung cancer cell line and CNE1 human NPC cell line were purchased from American Type Culture Collection. HeLa cells stably expressing GFP–LC3 and L929 cells stably expressing tfLC3 were kindly provided by Professor Han-ming Shen (National University of Singapore, Singapore). Cells were maintained in RPMI-1640 (Genom) supplemented with 10% FBS, 100 units/mL penicillin, 100 mg/mL streptomycin, 1 mM sodium pyruvate, and 1 mM nonessential amino acids. Cells were grown in 75-cm2 cell culture flasks (Biofil) at 37 °C in a 5% CO2 incubator (Thermo Scientific).
Cell viability assay
Cell viability was measured using the MTT assay. Cells (3 × 103) were seeded in 96-well plates and allowed to attach overnight in a 5% CO2 incubator. The cells were then treated with drugs at different concentrations. MTT (2.5 mg/mL) was added to each well and incubated for another 2 h at 37 °C. The supernatants were carefully aspirated, and the purple formazan crystals were dissolved in 200 µL of DMSO. The absorbance at 570 nm was measured with a Multiscan spectrophotometer (Thermo Scientific).
Colony formation assay
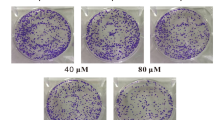
A total of 2 × 105 cells were seeded in a six-well plate for 18–24 h. After 48 h of GSE treatment, the original medium was removed, and the cells were resuspended in medium containing drugs at each concentration tested. Four hundred cells were seeded in triplicate in a six-well plate, incubated for another 9 days, photographed after staining with 0.1% (w/v) crystal violet and counted.
Reactive oxygen species (ROS) measurement
The intracellular ROS levels were measured using a ROS assay kit. A total of 2 × 105 cells were seeded into a six-well plate. After GSE treatment with or without NAC pretreatment, cells were incubated with DCFH-DA at 37 °C for 30 min. Hydrogen peroxide was used as a positive control. After trypsinization, fluorescence was measured on a flow cytometry (Beckman Coulter). The percent fluorescence intensity was used to represent the level of ROS generation.
Detection of cell apoptosis
Cell apoptosis was detected by staining using an Annexin V-FITC/PI apoptosis kit according to the manufacturer’s instructions. Briefly, A549 cells were digested with EDTA-free trypsin after GSE treatment and resuspended in 500 μL of binding buffer. After incubation with 5 μL of Annexin V-FITC and PI staining for 30 min at room temperature in the dark, data acquisition and analysis were carried out using flow cytometry.
Western blotting
Cells were seeded into 6-cm dishes (5 × 105 cells per dish) overnight and treated with GSE. After treatment, cells were collected by scraping and washed twice with PBS. Then, the cells were lysed with RIPA buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, sodium orthovanadate, sodium fluoride, EDTA, and leupeptin). Protein concentrations in the supernatants were determined with a BCA protein assay kit (Solarbio, PC0020). Equal amounts of proteins were separated on SDS-polyacrylamide gels, and then electroblotted onto polyvinylidene fluoride membranes (Bio-Rad, 1620184). The membranes were blocked with TBST plus 5% skimmed milk for 2 h and incubated with primary antibodies (1:1000) overnight at 4 °C. After washing three times with TBST, the membranes were incubated with secondary antibodies (1:5000) for 1 h at room temperature. Before development, the membranes were washed three times again, and the immunoblots were visualized with an ECL system. All the experiments were performed at least three times to ensure reproducibility.
Immunofluorescence
A549 cells were seeded into a chamber and treated with or without GSE. After treatment, cells were fixed with 4% formaldehyde in PBS for 20 min at 37 °C, permeabilized with 0.25% Triton X-100 in PBS for 15 min at room temperature, and then blocked with 1% BSA in PBS for 10 min. After that, the cells were incubated overnight with anti-Bax or anti-Bcl-2 antibody. After incubation with fluorescent secondary antibodies, the cells were observed under a confocal microscope.
Immunoprecipitation (IP)
HEK293 cells were seeded into 6-cm dishes overnight and then transfected with empty vector or vector for the expression of Myc-Bcl-2 plus Flag-Beclin 1. After 24 h, the cells were treated with GSE for 48 h. Finally, the cells were collected and lysed using IP lysis buffer (Thermo Scientific, 87787). Anti-Flag affinity gel (Sangon,D111139) was used to pull down proteins with a Flag tag overnight at 4 °C. After that, Western blotting was performed to determine the expression levels of Bcl-2.
Mitochondrial membrane potential (JC-1) assay
MMP (Δψm) was analyzed with an MMP assay kit with JC-1. Briefly, after the indicated treatments, cells were incubated with a JC-1 staining solution for 30 min at 37 °C. A Leica TCS SP8 microscope was used for observation and to take photographs.
Cell transfection with small interfering RNA
Briefly, cells were seeded into six-well plates overnight. Then, siRNA targeting Atg5 was transfected into A549 cells using Lipofectamine 2000 according to the manufacturer’s protocol. After 24 h, the cells were digested and reseeded into plates for experiments.
Subcellular localization
Fluorescence localization of subcellular structures (autophagosomes, lysosomes and the ER) in A549 cells was observed under a confocal microscope. An autophagosome formation assay was carried out in HeLa cells stably expressing GFP–LC3 or L929 cells expressing RFP-GFP–LC3.
Transmission electron microscopy
A549 cells were seeded into 5-cm petri dishes and treated with GSE. Then, the cells were collected with a scraper and centrifuged at 4 °C and 2000 rpm. The precipitated cells were fixed with 4% glutaraldehyde for another 2 h at room temperature and stored at 4 °C. Electron photomicrographs of the A549 cells were taken by Wuhan Goodbio Technology Co.
In vivo study
Female Balb/c nude mice (4–6 weeks old) were purchased from the Laboratory Animal Centre of the Zhejiang University of Technology (Hangzhou, China). Animal welfare was ensured, and the experimental procedures strictly followed the Guide for the Care and Use of Laboratory Animals. All efforts were made to minimize animal suffering and to reduce the number of animals used. Xenograft tumors were established by subcutaneously injecting 1.5 × 106 A549 cells in PBS in a total volume of 0.1 mL. After 1 week, to assess the effect of GSE on tumor growth, tumor-bearing mice were randomly divided into four groups: the vehicle group (10% DMSO, 20% polyethylene glycol, and 5% Tween-80 in PBS), GSE treatment group (40 mg/kg GSE), CQ treatment group (25 mg/kg CQ), and GSE plus CQ treatment group. All treatments were administered by intraperitoneal injection once every 2 days for 3 weeks.
Statistical analysis
All the experiments were performed at least three times in triplicate to ensure reproducibility. Differences among groups were analyzed by one-way variance, and the means of two groups were compared using Student’s t test with IBM Statistics SPSS 22. The results are expressed as the mean ± standard deviation. Differences with a **P value < 0.01 or *P value < 0.05 were considered statistically significant.
Results
GSE inhibited cancer cell proliferation and induced apoptosis in cancer cells
To evaluate the antiproliferative activity of GSE in cancer cells, several cell lines were exposed to GSE at different concentrations for 24, 48, and 72 h (Fig. 1b). In A549 and CNE1 cells, the cytotoxicity of GSE was shown to occur in a dose- and time-dependent manner (Fig. 1b). In addition, we determined the effect of GSE on cell proliferation by colony formation assay. The results showed that GSE significantly decreased colony formation in both A549 and CNE1 cells (Fig. 1c, d).
To elucidate the type of cell death caused by GSE, we analyzed apoptosis markers by flow cytometry and Western blotting. The percentages of apoptotic cells in the GSE-treated group and the untreated group were 35.4% and 2.5% (Fig. 1e), respectively. Western blotting results also demonstrated that the cleavage of caspase 3 was increased after GSE treatment in both A549 and CNE1 cells (Fig. 1f). Moreover, immunofluorescence was performed to examine the expression levels of Bax and Bcl-2, and the ratio of Bcl-2 to Bax was significantly decreased by GSE (Fig. 1g, h), further confirming that GSE induced cell apoptosis.
GSE increased autophagic flux in cancer cells
To ascertain the induction of autophagy, cells were treated with GSE, and the autophagosome marker microtubule-associated protein 1 LC3 was assessed. As shown, the number of GFP–LC3 puncta, indicating the formation of autophagosomes, was significantly increased by GSE (Fig. 2a, b). Moreover, assessment of the ultrastructure of A549 cells showed the presence of double-membrane autophagosomes in GSE-treated cells (Fig. 2c). In addition, the protein levels of LC3 were dramatically increased by GSE, as shown by Western blot analysis (Fig. 2d). For the purpose of this article, autophagy induction specifically refers to an increase in autophagic flux rather than simply an increase in autophagic markers in cells. Thus, we detected changes in autophagy flux using the lysosomal inhibitor CQ and observed a further increase in LC3 levels induced by GSE in both A549 and CNE1 cells (Fig. 2e). This result suggests that autophagy flux was also enhanced under GSE treatment.
a, b HeLa cells stably expressing GFP–LC3 were treated with GSE for 48 h. Confocal images of GFP–LC3 puncta are shown (scale bar: 25 µm). GFP puncta were quantified by ImageJ. **P < 0.01. c TEM images show the ultrastructure of A549 cells under GSE treatment (scale bar 0.2 μm). ▲ indicates double-membrane autophagosomes. d Expression of the LC3 protein in A549 and CNE1 cells under GSE treatment was determined. e Expression of the LC3 protein with or without CQ pretreatment in A549 and CNE1 cells under GSE treatment for 48 h was determined. f The interaction between Bcl-2 and Beclin 1 was determined using IP. GAPDH was used as a loading control. g, h LysoTracker staining was performed in GSE-treated A549 cells examined under confocal microscopy(scale bar: 50 µm), and cell fluorescence was measured by flow cytometry. *P < 0.05. i, j HeLa–GFP–LC3 cells were treated with GSE in the presence of the autophagy inhibitor CQ or the autophagy inducer rapamycin, and then stained with LysoTracker. The colocalization of lysosomes and GFP–LC3 was determined under a confocal microscope. The colocalization coefficient was detected using Image-Pro Plus. *P < 0.05. k, l L929 cells stably expressing fLC3B were treated with GSE with or without an autophagy inhibitor or inducer. Confocal images of GFP–LC3 and RFP–LC3 puncta are shown (scale bar: 25 µm). *P < 0.05.
To investigate the relationship between autophagy and apoptosis induced by GSE, IP was performed to assess the interaction between Beclin 1 and Bcl-2. Beclin 1, an antiapoptotic protein that interacts with Bcl-2, is important in autophagy induction. We first transfected HEK293 cells with plasmids for the expression of Myc-Bcl-2 and Flag-Beclin 1, and then treated the cells with GSE. As shown in Fig. 2f, GSE treatment attenuated the interaction between Beclin 1 and Bcl-2, allowing the dissociation of Beclin 1 from Bcl-2 and the subsequent stimulation of autophagy.
To examine the effect of GSE on lysosomal function, LysoTracker staining was performed, and the fluorescence intensity of the cells was measured by flow cytometry and confocal microscopy. As shown, a significant increase in cell fluorescence (Fig. 2g, h) and the colocalization of lysosomes and GFP–LC3 (Fig. 2i, j) were observed, indicating enhanced lysosomal acidification and the formation of autolysosomes. In addition, we used mRFP-GFP tandem fluorescent‑tagged LC3B expression in cells to examine autophagosome–lysosome fusion. In cells transfected with this RFP-GFP tandem construct, the RFP component is stable under the acidic environment of the lysosome, while GFP is degraded. Thus, the number of RFP-positive puncta can be used as an indicator of autolysosome formation. As shown in Fig. 2k, l, we observed significant increases in puncta positive for only RFP and RFP- and GFP-positive puncta following GSE treatment, confirming the enhanced formation of autolysosomes.
GSE treatment activated endoplasmic reticulum (ER) stress
We previously observed that GSE treatment could induce the cellular vacuolization of A549 and CNE1 cells under light microscopy by Giemsa staining (Fig. 3a). To investigate this phenomenon, we stained GSE-treated cells with ER Tracker and found vacuoles around the ER under confocal microscopy, indicating the presence of ER stress (Fig. 3b). The ER has an important function in maintaining calcium homeostasis, and calcium disorder results in ER stress [26]. In this study, Fluo-3 AM was used to label cytoplasmic calcium in A549 cells. Under confocal microscopy, a significantly elevated cytoplasmic calcium level was detected in GSE-treated cells (Fig. 3c, d). Consistently, the expression levels of the UPR markers GRP78, IRE1α, and ATF4 were also increased by GSE treatment (Fig. 3e). These findings strongly demonstrate that GSE induced ER stress in A549 cells.
a Vacuoles in A549 and CNE1 cells following the indicated GSE treatment were observed under a light microscope (scale bar: 20 μm). b ER localization in GSE-treated A549 cells stained using ER Tracker (scale bar: 25 µm) was determined with confocal images. c, d The colocalization of ER and Ca2+ in GSE-treated A549 cells was determined using confocal microscopy (scale bar: 25 µm). The colocalization coefficient was detected using Image-Pro Plus. **P < 0.01. e The expression of ER stress-related proteins in cells under GSE treatment was measured using Western blotting. GAPDH was used as a loading control. f The expression of autophagy-related proteins was determined in GSE-treated cells after the knockdown of IRE1α. GAPDH was used as a loading control. g, h The formation of GFP–LC3 puncta in GSE-treated cells in which IRE1α was knocked down was assessed by confocal microscopy (scale bar: 25 µm). GFP puncta were quantified by ImageJ. *P < 0.05. i The expression of autophagy-related proteins was determined in GSE-treated cells in which IRE1α was inhibited by 4μ8c.
GSE induced autophagy through the activation of ER stress
ER stress is closely related to the activation of autophagy, an important and evolutionarily conserved mechanism for the maintenance of cellular homeostasis [26]. Functionally, autophagy is a double-edged sword in cancer, as it can either promote or suppress cancer cell growth in the tumor microenvironment. To further assess the role of ER stress in autophagy induction, we knocked down IRE1α in A549 cells and found that the increase in the autophagy marker LC3 by GSE was impaired in IRE1α-knockdown cells (Fig. 3f). Consistently, the increase in GFP–LC3 puncta induced by GSE was also attenuated when IRE1α was knocked down (Fig. 3g, h), indicating the involvement of ER stress in autophagy induction. In addition, the IRE1α inhibitor 4μ8c was employed to block the IRE1α pathway. As shown in Fig. 3i, we observed the opposing effects of 4μ8c and GSE on autophagy induction. These data demonstrated that ER stress is required for GSE induced autophagy.
GSE induces ER stress through ROS generation
To reveal the mechanism by which GSE induces ER stress, we measured ROS production using the fluorescent probe DCFH-DA. As shown, GSE significantly increased ROS generation in a dose-dependent manner in A549 cells (Fig. 4a). The main source of cellular ROS is mitochondria. Under GSE treatment, we observed a decrease in MMP (Fig. 4b), accompanied by a decrease in red fluorescence (aggregated JC-1) and an increase in green fluorescence (monomeric JC-1). Consequently, these changes in the Bcl-2 protein family illustrated mitochondrial dysfunction (Fig. 4c).
a ROS generation in A549 cells under the indicated GSE treatment was determined using DCFH-DA staining. The results were quantified using flow cytometry and statistically analyzed. *P < 0.05, **P < 0.01. b Confocal images of JC-1-stained GSE-treated cells are shown (scale bar: 25 µm). c The expression of mitochondria-related apoptotic proteins was assessed in A549 cells under GSE treatment. GAPDH was used as a loading control. d Vacuoles in GSE-treated A549 cells with or without NAC treatment were compared under a light microscope (scale bar: 20 μm). e ROS generation in GSE-treated A549 cells pretreated with NAC was determined using the probe DCFH-DA. The results were quantified using flow cytometry and statistically analyzed. **P < 0.01. f GRP78 expression levels were assessed in GSE-treated A549 cells with or without NAC pretreatment. GAPDH was used as a loading control. g, h Colocalization of the ER and Ca2+ in A549 cells treated with GSE in the presence or absence of NAC was determined with confocal microscopy (scale bar: 25 µm). The colocalization coefficient was detected using Image-Pro Plus. *P < 0.05. i Caspase 3 expression levels in A549 cells with GSE treatment in the presence or absence of NAC were determined. GAPDH was used as a loading control. j Cell viability in GSE-treated A549 cells in which ROS were scavenged with NAC was determined using the MTT assay. **P < 0.01. k The A549 cell apoptosis rate was determined using Annexin V-FITC/PI staining coupled with flow cytometry. **P < 0.01.
Next, we determined the role of ROS production in ER stress activated by GSE. With pretreatment with the ROS scavenger NAC, the degree of vacuolation in GSE-treated cells observed under a light microscope was significantly decreased (Fig. 4d), in accordance with the decrease in ROS levels (Fig. 4e). This result indicated that GSE-activated ER stress may have been alleviated. In addition, we determined the protein levels of UPR markers and found that NAC pretreatment attenuated the increase in GRP78 induced by GSE (Fig. 4f). Under confocal microscopy, ER Tracker and Fluo-3 AM staining showed that the increase in cytoplasmic calcium levels induced by GSE was also impaired (Fig. 4g, h), suggesting that the release of Ca2+ was partially inhibited by NAC. The above results demonstrate that ROS generation is a key initiator of ER stress induced by GSE.
Finally, we investigated whether ROS production was involved in GSE-induced cell apoptosis. Thus, we examined the protein levels of caspase 3 following treatment with GSE and the ROS scavenger NAC. As shown in Fig. 4i, the cleavage of caspase 3 induced by GSE was partially suppressed by NAC pretreatment, suggesting the inductive effect of ROS accumulation on apoptosis. Consistently, decreases in the cell death and apoptosis rates induced by NAC pretreatment also demonstrated that ROS production leads to cell apoptosis induced by GSE (Fig. 4j, k).
Autophagy is involved in GSE-induced cancer cell death
To reveal the role of autophagy in the cytotoxicity of GSE, we first assessed the viability of A549 cells under treatment with GSE plus chloroquine (CQ, an autophagy inhibitor). Compared with GSE treatment alone, the presence of CQ significantly increased cell viability (Fig. 5a), indicating that autophagy served as the type of cell death induced by GSE. Next, we knocked down Atg5, a key autophagy-related gene, in A549 cells and observed the morphological changes in these A549 cells under GSE treatment. As shown, knockdown of Atg5 reversed the cell death induced by GSE treatment observed under a light microscope (Fig. 5b). Consistently, the apoptosis rate was also lower in Atg5-knockdown cells under GSE treatment compared to control cells (Fig. 5c). Meanwhile, the increase in cleaved caspase 3 levels induced by GSE was also attenuated when Atg5 was knocked down (Fig. 5d), clarifying the role of autophagy in promoting apoptosis. Taken together, these data demonstrate that autophagy is required for the cancer cell death caused by GSE and that GSE exerts an anticancer effect through autophagy.
a A549 cell viabilities under GSE treatment with or without the autophagy inhibitor CQ were determined using the MTT assay and compared. **P < 0.01. b Changes in cell morphology in Atg5-knockdown A549 cells treated with GSE were determined under a light microscope (scale bar: 50 μm). c The cell apoptosis rate in GSE-treated A549 cells with Atg5 knockdown was determined using Annexin V-FITC/PI staining coupled with flow cytometry. *P < 0.05, **P < 0.01. d The expression of cleaved caspase 3 in Atg5-knockdown A549 cells under GSE treatment was determined by Western blotting. GAPDH was used as a loading control. e The effect of GSE treatment on tumor growth in mice was measured by tumor volume. *P < 0.05, **P < 0.01. f Tumors were taken from tumor-bearing mice after sacrifice and photographed. g Tumor weight was measured and statistically analyzed. *P < 0.05. h Schematic diagram of the anticancer mechanism of GSE.
GSE exerts an antitumor effect in vivo through autophagy induction
To evaluate the antitumor effect of GSE in vivo, xenograft model mice were intraperitoneally treated with GSE. The autophagy inhibitor CQ was used to determine the role of autophagy in tumor suppression. As shown in Fig. 5e, treatment with 40 mg/kg GSE inhibited tumor xenograft growth by 72.8%, but cotreatment with 25 mg/kg CQ inhibited tumor xenograft growth by only 18.3%. This result indicates that GSE exerts an autophagy-mediated tumor-suppressive effect. No mice died during the experiment, and no significant difference in body weight between the control and treatment groups was observed (data not shown). After the mice were sacrificed, the tumors were resected, and the tumors were weighed. A significant decrease in tumor weight was observed in the GSE treatment group, but this decrease was reversed in the presence of CQ (Fig. 5f, g). The above results demonstrated that GSE exerts an antitumor effect through autophagy, which is consistent with our in vitro results.
Discussion
Morus alba L. is a traditional Chinese medicine widely applied for the treatment of respiratory diseases. More recently, it was proven[27] that compounds extracted from Morus alba L. could be beneficial in the treatment of human cancers. For example, morusin could induce apoptosis and suppress NF-κB activity in cancer cells [28]. GSE is a secondary metabolite extracted from the leaves of Morus alba L. Here, we conducted a novel study on the anticancer activity of GSE in human respiratory cancer. In this study, we have shown for the first time that GSE induced autophagy-mediated cell death via ER stress and that this effect was mediated by ROS generation (Fig. 5h). Cell apoptosis was also triggered by ROS generation under GSE treatment. The accumulation of ROS caused the UPR and induced autophagy. Interestingly, inhibition of autophagy reduced GSE-induced cell death, indicating that autophagy plays a critical role in cell apoptosis.
Autophagy is a highly regulated process that serves as a dynamic system by which cellular proteins and organelles are recycled to produce new building blocks and energy for cellular renovation and homeostasis [29]. The autophagosome, the intermediate organelle in the autophagy process, sequesters a small portion of the cytoplasm into an isolated membrane-bound vesicle. It fuses with the lysosome to form an autolysosome in which the molecules and organelles contained within are degraded [30]. In our study, the formation of GFP–LC3 puncta and double-membrane autophagosomes showed that GSE induced autophagy in A549 and CNE1 cells (Fig. 2a–c). We also used the autophagy inhibitor CQ to evaluate autophagy flux, which showed a further increase in LC3 protein levels under treatment with GSE plus CQ, suggesting that autophagy flux was also enhanced (Fig. 2e). This could be attributed to enhanced lysosomal activity (Fig. 2g, h) and the fusion of lysosomes and autophagosomes (Fig. 2i–l), demonstrating the positive effect of GSE on the autophagy process.
Generally, inhibition of autophagy attenuates the ability of cells to overcome stress and maintain homeostasis. However, given that autophagy is a cellular response to various stresses, such as starvation or inflammation, it is not surprising that cells also show features of autophagy as they die [31]. However, autophagy neither promotes nor inhibits apoptosis in lung cancer cells [32]. Recently, many studies have reported the prodeath role of autophagy in anticancer processes, and signs of autophagy in dying cells were interpreted as indicating the use of autophagy as a death mechanism [32,33,34,35,36]. In our study, the autophagy inhibitor CQ and small interfering RNA were used to explore the role of autophagy in the process of cell death. Cell death induced by GSE was decreased in the presence of CQ (Fig. 5a) or knockdown of Atg5 (Fig. 5b–d), which supports the notion that autophagy functions to promote cell death. The crosstalk between autophagy and cell death suggests that the role of autophagy may be context specific, and revealing their relationship will aid in understanding the contribution of autophagy to cell death [37].
The most studied mechanism employed in the ER is the UPR, which is known to cause a series of effects, such as autophagy [38]. Three conserved ER transmembrane proteins involved in ER stress, IRE1α, activating transcription factor 6 and pancreatic ER kinase-like ER kinase, bind globulin protein (also called GRP78 or Kar2p) under normal conditions [38]. The ER is also involved in intracellular Ca2+ homeostasis. Following ER stress, calcium is released from the lumen to the cytosol through Ca2+ channels; this increase in cytoplasmic calcium causes abnormal MMP and activates prodeath autophagy [39, 40]. For example, the natural product cryptotanshinone induced prodeath autophagy in lung cancer [36]. In our study, the release of Ca2+ from the ER and the upregulation of GRP78 expression indicated the occurrence of ER stress. Ca2+ was found to be released, and the IRE1α pathway was activated after GSE treatment (Fig. 3c–e). The partial decrease in autophagy when ER stress was inhibited indicated the positive regulatory effect of GSE treatment on autophagy (Fig. 3f–i).
ROS are a series of crucial signaling molecules in the oxidative stress response. The excessive accumulation of ROS results in oxidative stress and mitochondrial dysfunction [41], which further leads to autophagy and cell injury [42]. Recent studies have shown that ROS act as cellular signaling molecules and initiate autophagosome formation and autophagic degradation [43]. Conversely, autophagy serves to reduce oxidative damage through the removal of protein aggregates and damaged organelles, such as mitochondria [44]. The UPR is carried out through a collection of adaptive signaling pathways that resolve protein misfolding and restore an efficient protein-folding environment. Many studies suggest that altered redox homeostasis in the ER is sufficient to cause ER stress, which in turn induces the production of ROS in the ER and mitochondria [45]. In this study, we observed the accumulation of ROS and a decrease in MMP under GSE treatment (Fig. 4a, b). We found that when ROS were scavenged, ER stress, and autophagy activated by GSE were also attenuated accordingly. Therefore, we conclude that ROS constitute a pathway upstream of ER stress and autophagy. At the same time, ROS scavenging also decreased cell death (Fig. 4i–k). The above findings reveal the anticancer mechanism of GSE in lung cancer. We will explore the anticancer activity of GSE in vivo in the future.
In summary, the effects of GSE on autophagy and apoptosis provide a new research field in the treatment of respiratory cancers and a new possibility towards the development of cancer therapeutics with traditional Chinese medicine. Great progress will be achieved through the development of more effective drugs, leading to a promising therapy.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108.
Goldstraw P, Ball D, Jett JR, Le Chevalier T, Lim E, Nicholson AG, et al. Non-small-cell lung cancer. Lancet. 2011;378:1727–40.
Remark R, Becker C, Gomez JE, Damotte D, Dieu-Nosjean MC, Sautes-Fridman C, et al. The non-small cell lung cancer immune contexture. A major determinant of tumor characteristics and patient outcome. Am J Respir Crit Care Med. 2015;191:377–90.
Henschke CI, Yip R, Smith JP, Wolf AS, Flores RM, Liang M, et al. CT screening for lung cancer: part-solid nodules in baseline and annual repeat rounds. Am J Roentgenol. 2016;207:1176–84.
Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311.
Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–54.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Syah YM, Achmad SA, Ghisalberti EL, Hakim EH, Iman MZ, Makmur L, et al. Andalasin A, a new stilbene dimer from Morus macroura. Fitoterapia. 2000;71:630–5.
Yang Y, Tan YX, Chen RY, Kang J. The latest review on the polyphenols and their bioactivities of Chinese Morus plants. J Asian Nat Prod Res. 2014;16:690–702.
Wei H, Zhu J-j, Liu X-q, Feng W-h, Wang Z-m, Yan L-H, et al. Review of bioactive compounds from root barks of Morus plants (Sang-Bai-Pi) and their pharmacological effects. Cogent Chem. 2016;2:1212320.
Shuang E, Yamamoto K, Sakamoto Y, Mizowaki Y, Iwagaki Y, Kimura T, et al. Intake of mulberry 1-deoxynojirimycin prevents colorectal cancer in mice. J Clin Biochem Nutr. 2017;61:47–52.
Min TR, Park HJ, Park MN, Kim B, Park SH. The root bark of Morus alba L. suppressed the migration of human non-small-cell lung cancer cells through inhibition of epithelial(-)mesenchymal transition mediated by STAT3 and Src. Int J Mol Sci. 2019;20:2244.
Cho SW, Na W, Choi M, Kang SJ, Lee SG, Choi CY. Autophagy inhibits cell death induced by the anti-cancer drug morusin. Am J Cancer Res. 2017;7:518–30.
Guo F, Zou Y, Zheng Y. Moracin M inhibits lipopolysaccharide-induced inflammatory responses in nucleus pulposus cells via regulating PI3K/Akt/mTOR phosphorylation. Int Immunopharmacol. 2018;58:80–6.
Gu Y, Gao L, Chen Y, Xu Z, Yu K, Zhang D, et al. Sanggenon C protects against cardiomyocyte hypoxia injury by increasing autophagy. Mol Med Rep. 2017;16:8130–6.
Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186:773–82.
Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12 (Suppl 2):1542–52.
Murrow L, Debnath J. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu Rev Pathol. 2013;8:105–37.
Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:859–64.
Nihira K, Miki Y, Iida S, Narumi S, Ono K, Iwabuchi E, et al. An activation of LC3A-mediated autophagy contributes to de novo and acquired resistance to EGFR tyrosine kinase inhibitors in lung adenocarcinoma. J Pathol. 2014;234:277–88.
Platania M, Agustoni F, Formisano B, Vitali M, Ducceschi M, Pietrantonio F, et al. Clinical retrospective analysis of erlotinib in the treatment of elderly patients with advanced non-small cell lung cancer. Target Oncol. 2011;6:181–6.
Lee JG, Wu R. Combination erlotinib-cisplatin and Atg3-mediated autophagy in erlotinib resistant lung cancer. PLoS One. 2012;7:e48532.
Sun X, Yan P, Zou C, Wong Y-K, Shu Y, Lee YM, et al. Targeting autophagy enhances the anticancer effect of artemisinin and its derivatives. Med Res Rev. 2019;39:2172–93.
Gorzalczany Y, Gilad Y, Amihai D, Hammel I, Sagi-Eisenberg R, Merimsky O. Combining an EGFR directed tyrosine kinase inhibitor with autophagy-inducing drugs: a beneficial strategy to combat non-small cell lung cancer. Cancer Lett. 2011;310:207–15.
Chaabane W, User SD, El-Gazzah M, Jaksik R, Sajjadi E, Rzeszowska-Wolny J, et al. Autophagy, apoptosis, mitoptosis and necrosis: interdependence between those pathways and effects on cancer. Arch Immunol Ther Exp. 2013;61:43–58.
Senft D, Ronai ZA. UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem Sci. 2015;40:141–8.
Yu JS, Lee D, Lee SR, Lee JW, Choi CI, Jang TS, et al. Chemical characterization of cytotoxic indole acetic acid derivative from mulberry fruit (Morus alba L.) against human cervical cancer. Bioorg Chem. 2018;76:28–36.
Lee JC, Won SJ, Chao CL, Wu FL, Liu HS, Ling P, et al. Morusin induces apoptosis and suppresses NF-kappaB activity in human colorectal cancer HT-29 cells. Biochem Biophys Res Commun. 2008;372:236–42.
Onorati AV, Dyczynski M, Ojha R, Amaravadi RK. Targeting autophagy in cancer. Cancer. 2018;124:3307–18.
Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–41.
Denton D, Nicolson S, Kumar S. Cell death by autophagy: facts and apparent artefacts. Cell Death Differ. 2012;19:87–95.
Tang ZH, Chen X, Wang ZY, Chai K, Wang YF, Xu XH, et al. Induction of C/EBP homologous protein-mediated apoptosis and autophagy by licochalcone A in non-small cell lung cancer cells. Sci Rep. 2016;6:26241.
Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH, et al. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr Biol. 2009;19:1741–6.
Liu Y, Shoji-Kawata S, Sumpter RM Jr., Wei Y, Ginet V, Zhang L, et al. Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc Natl Acad Sci USA. 2013;110:20364–71.
Dang S, Yu ZM, Zhang CY, Zheng J, Li KL, Wu Y, et al. Autophagy promotes apoptosis of mesenchymal stem cells under inflammatory microenvironment. Stem Cell Res Ther. 2015;6:247.
Zhang M, Su L, Xiao Z, Liu X, Liu X. Methyl jasmonate induces apoptosis and pro-apoptotic autophagy via the ROS pathway in human non-small cell lung cancer. Am J Cancer Res. 2016;6:187–99.
Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–93.
Mori K. The unfolded protein response: the dawn of a new field. Proc Jpn Acad B Phys. 2015;91:469–80.
Maheswari U, Ghosh K, Sadras SR. Licarin A induces cell death by activation of autophagy and apoptosis in non-small cell lung cancer cells. Apoptosis. 2018;23:210–25.
Liu G, Pei F, Yang F, Li L, Amin AD, Liu S, et al. Role of autophagy and apoptosis in non-small-cell lung cancer. Int J Mol Sci. 2017;18:367.
Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBOJ. 2007;26:1749–60.
Fandy TE, Jiemjit A, Thakar M, Rhoden P, Suarez L, Gore SD. Decitabine induces delayed reactive oxygen species (ROS) accumulation in leukemia cells and induces the expression of ROS generating enzymes. Clin Cancer Res. 2014;20:1249–58.
Chen Y, Azad MB, Gibson SB. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009;16:1040–52.
Ureshino RP, Rocha KK, Lopes GS, Bincoletto C, Smaili SS. Calcium signaling alterations, oxidative stress, and autophagy in aging. Antioxid Redox Signal. 2014;21:123–37.
Cao SS, Kaufman RJ. Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxid Redox Signal. 2014;21:396–413.
Acknowledgements
This work was supported by the National Science and Technology Major Project of China (2019ZX09301004), Zhejiang Provincial Science and Technology Planning Project (2016C04005) to JKT; Zhejiang Provincial Outstanding Talent Project of Ten Thousand Talents Program, Zhejiang Provincial Qianjiang Talents Program, Zhejiang Provincial Natural Science Foundation (LR18H160002), Traditional Chinese Medicine Scientific Research Fund Project of Zhejiang Province (2018ZB010), Zhejiang Provincial Program in Medicine and Health Sciences and Technology (2018253645) to JBZ; National Natural Science Foundation of China (81703907) to XS; Zhejiang Provincial Program in Medicine and Health Sciences and Technology (2018KY223) to SFY.
Author information
Authors and Affiliations
Contributions
JBZ and JKT designed research; HHY, YHS, CCG, and MTX performed research; RLG and SFY contributed new reagents or analytic tools; YTH, XS, and SXL analyzed data; HHY and YHS wrote the paper; HTH and ZPW drafted the rebutal letter.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
About this article
Cite this article
Shu, Yh., Yuan, Hh., Xu, Mt. et al. A novel Diels–Alder adduct of mulberry leaves exerts anticancer effect through autophagy-mediated cell death. Acta Pharmacol Sin 42, 780–790 (2021). https://doi.org/10.1038/s41401-020-0492-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-020-0492-5
Keywords
This article is cited by
-
The evolutionary origin of naturally occurring intermolecular Diels-Alderases from Morus alba
Nature Communications (2024)
-
Mulberry Diels–Alder-type adducts: isolation, structure, bioactivity, and synthesis
Natural Products and Bioprospecting (2022)
-
Biological activities of Egyptian grape and mulberry by-products and their potential use as natural sources of food additives and nutraceuticals foods
Journal of Food Measurement and Characterization (2022)