Abstract
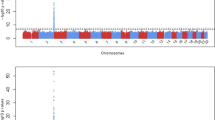
We evaluated interactions of SNP-by-ACE-I/ARB and SNP-by-TD on serum potassium (K+) among users of antihypertensive treatments (anti-HTN). Our study included seven European-ancestry (EA) (N = 4835) and four African-ancestry (AA) cohorts (N = 2016). We performed race-stratified, fixed-effect, inverse-variance-weighted meta-analyses of 2.5 million SNP-by-drug interaction estimates; race-combined meta-analysis; and trans-ethnic fine-mapping. Among EAs, we identified 11 significant SNPs (P < 5 × 10−8) for SNP-ACE-I/ARB interactions on serum K+ that were located between NR2F1-AS1 and ARRDC3-AS1 on chromosome 5 (top SNP rs6878413 P = 1.7 × 10−8; ratio of serum K+ in ACE-I/ARB exposed compared to unexposed is 1.0476, 1.0280, 1.0088 for the TT, AT, and AA genotypes, respectively). Trans-ethnic fine mapping identified the same group of SNPs on chromosome 5 as genome-wide significant for the ACE-I/ARB analysis. In conclusion, SNP-by-ACE-I /ARB interaction analyses uncovered loci that, if replicated, could have future implications for the prevention of arrhythmias due to anti-HTN treatment-related hyperkalemia. Before these loci can be identified as clinically relevant, future validation studies of equal or greater size in comparison to our discovery effort are needed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Papademetriou V. Diuretics, hypokalemia, and cardiac arrhythmia: a 20-year controversy. J Clin Hypertens. 2006;8:86–92.
Cohen HW, Madhavan S, Alderman MH. High and low serum potassium associated with cardiovascular events in diuretic-treated patients. J Hypertens. 2001;19:1315–23.
Salvetti A, Ghiadoni L. Thiazide diuretics in the treatment of hypertension: an update. J Am Soc Nephrol. 2006;17:S25–9.
Freis ED. The efficacy and safety of diuretics in treating hypertension. Ann Intern Med. 1995;122:223–6.
Franse LV, Pahor M, Di Bari M, Somes GW, Cushman WC, Applegate WB. Hypokalemia associated with diuretic use and cardiovascular events in the Systolic Hypertension in the Elderly Program. Hypertension. 2000;35:1025–30.
Alderman MH, Piller LB, Ford CE, Probstfield JL, Oparil S, Cushman WC, et al. Clinical significance of incident hypokalemia and hyperkalemia in treated hypertensive patients in the antihypertensive and lipid-lowering treatment to prevent heart attack trial. Hypertension. 2012;59:926–33.
Bathum L, Fagnani C, Christiansen L, Christensen K. Heritability of biochemical kidney markers and relation to survival in the elderly–results from a Danish population-based twin study. Clin Chim Acta. 2004;349:143–50.
Marroni F, Grazio D, Pattaro C, Devoto M, Pramstaller P. Estimates of genetic and environmental contribution to 43 quantitative traits support sharing of a homogeneous environment in an isolated population from South Tyrol, Italy. Hum Hered. 2008;65:175–82.
Nilsson SE, Read S, Berg S, Johansson B. Heritabilities for fifteen routine biochemical values: findings in 215 Swedish twin pairs 82 years of age or older. Scand J Clin Lab Invest. 2009;69:562–9.
Pilia G, Chen WM, Scuteri A, Orru M, Albai G, Dei M, et al. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2006;2:e132.
Psaty BM, O’Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2:73–80.
Li Y, Abecasis GR. MACH 1.0: rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet. 2006;S79:2290.
Browning BL, Yu Z. Simultaneous genotype calling and haplotype phasing improves genotype accuracy and reduces false-positive associations for genome-wide association studies. Am J Hum Genet. 2009;85:847–61.
Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3:e114.
Voorman A, Lumley T, McKnight B, Rice K. Behavior of QQ-plots and genomic control in studies of gene-environment interaction. PLoS ONE. 2011;6:e19416.
Chang AR, Sang Y, Leddy J, Yahya T, Kirchner HL, Inker LA, et al. Antihypertensive medications and the prevalence of hyperkalemia in a large health system. Hypertension. 2016;67:1181–8.
Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1.
Sitlani CM, Rice KM, Lumley T, McKnight B, Cupples LA, Avery CL, et al. Generalized estimating equations for genome-wide association studies using longitudinal phenotype data. Stat Med. 2015;34:118–30.
Satterthwaite FE. An approximate distribution of estimates of variance components. Biometrics. 1946;2:110–4.
Pan W, Wall MM. Small-sample adjustments in using the sandwich variance estimator in generalized estimating equations. Stat Med. 2002;21:1429–41.
Morris AP. Transethnic meta-analysis of genomewide association studies. Genet Epidemiol. 2011;35:809–22.
Wang X, Chua HX, Chen P, Ong RT, Sim X, Zhang W, et al. Comparing methods for performing trans-ethnic meta-analysis of genome-wide association studies. Hum Mol Genet. 2013;22:2303–11.
Coetzee SG, Rhie SK, Berman BP, Coetzee GA, Noushmehr H. FunciSNP: an R/bioconductor tool integrating functional non-coding data sets with genetic association studies to identify candidate regulatory SNPs. Nucleic Acids Res. 2012;40:e139.
Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–7.
Rafiq S, Khan S, Tapper W, Collins A, Upstill-Goddard R, Gerty S, et al. A genome wide meta-analysis study for identification of common variation associated with breast cancer prognosis. PLoS ONE. 2014;9:e101488.
Fox CS, Liu Y, White CC, Feitosa M, Smith AV, Heard-Costa N, et al. Genome-wide association for abdominal subcutaneous and visceral adipose reveals a novel locus for visceral fat in women. PLoS Genet. 2012;8:e1002695.
Comuzzie AG, Cole SA, Laston SL, Voruganti VS, Haack K, Gibbs RA, et al. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS ONE. 2012;7:e51954.
Daily K, Patel VR, Rigor P, Xie X, Baldi P. MotifMap: integrative genome-wide maps of regulatory motif sites for model species. BMC Bioinf. 2011;12:495.
Qi S, O’Hayre M, Gutkind JS, Hurley JH. Insights into beta2-adrenergic receptor binding from structures of the N-terminal lobe of ARRDC3. Protein Sci: a Publ Protein Soc. 2014;23:1708–16.
Moratinos J, Reverte M. Effects of catecholamines on plasma potassium: the role of alpha- and beta-adrenoceptors. Fundam Clin Pharmacol. 1993;7:143–53.
Haghikia A, Ricke-Hoch M, Stapel B, Gorst I, Hilfiker-Kleiner D. STAT3, a key regulator of cell-to-cell communication in the heart. Cardiovasc Res. 2014;102:281–9.
Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–87.
Fox ER, Young JH, Li Y, Dreisbach AW, Keating BJ, Musani SK, et al. Association of genetic variation with systolic and diastolic blood pressure among African Americans: the Candidate Gene Association Resource study. Hum Mol Genet. 2011;20:2273–84.
Frank DU, Carter KL, Thomas KR, Burr RM, Bakker ML, Coetzee WA, et al. Lethal arrhythmias in Tbx3-deficient mice reveal extreme dosage sensitivity of cardiac conduction system function and homeostasis. Proc Natl Acad Sci USA. 2012;109:E154–63.
Vandell AG, McDonough CW, Gong Y, Langaee TY, Lucas AM, Chapman AB, et al. Hydrochlorothiazide-induced hyperuricaemia in the pharmacogenomic evaluation of antihypertensive responses study. J Intern Med. 2014;276:486–97.
Del-Aguila JL, Beitelshees AL, Cooper-Dehoff RM, Chapman AB, Gums JG, Bailey K, et al. Genome-wide association analyses suggest NELL1 influences adverse metabolic response to HCTZ in African Americans. Pharm J. 2014;14:35–40.
Del-Aguila JL, Cooper-DeHoff RM, Chapman AB, Gums JG, Beitelshees AL, Bailey K, et al. Transethnic meta-analysis suggests genetic variation in the HEME pathway influences potassium response in patients treated with hydrochlorothiazide. Pharm J. 2015;15:153–7.
Huang CC, Chung CM, Hung SI, Leu HB, Lin LY, Huang PH, et al. Genetic predictors of thiazide-induced serum potassium changes in nondiabetic hypertensive patients. Hypertens Res. 2014;37:759–64.
Button KS, Ioannidis JP, Mokrysz C, Nosek BA, Flint J, Robinson ES, et al. Power failure: why small sample size undermines the reliability of neuroscience. Nat Rev Neurosci. 2013;14:365–76.
Ashton JC. Experimental power comes from powerful theories–the real problem in null hypothesis testing. Nat Rev Neurosci. 2013;14:585.
Quinlan PT. Misuse of power: in defence of small-scale science. Nat Rev Neurosci. 2013;14:585.
Bacchetti P. Small sample size is not the real problem. Nat Rev Neurosci. 2013;14:585.
Button KS, Ioannidis JP, Mokrysz C, Nosek BA, Flint J, Robinson ES, et al. Confidence and precision increase with high statistical power. Nat Rev Neurosci. 2013;14:585–6.
Kraft P, Zeggini E, Ioannidis JP. Replication in genome-wide association studies. Stat Sci. 2009;24:561–73.
Funding
Cardiovascular Health Study: This CHS research was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086; and NHLBI grants U01HL080295, R01HL087652, R01HL105756, R01HL103612, R01HL120393, and R01HL130114 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through R01AG023629 from the National Institute on Aging (NIA). A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center.
Jackson Heart Study: We thank the Jackson Heart Study (JHS) participants and staff for their contributions to this work. The JHS is supported by contracts HHSN268201300046C, HHSN268201300047C, HHSN268201300048C, HHSN268201300049C, HHSN268201300050C from the National Heart, Lung, and Blood Institute and the National Institute on Minority Health and Health Disparities.
Rotterdam Study: The RS is supported by the Erasmus Medical Center and Erasmus University Rotterdam; The Netherlands Organization for Scientific Research; The Netherlands Organization for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly; The Netherlands Heart Foundation; the Ministry of Education, Culture and Science; the Ministry of Health Welfare and Sports; the European Commission; and the Municipality of Rotterdam. Support for genotyping was provided by The Netherlands Organization for Scientific Research (NWO) (175.010.2005.011, 911.03.012) and Research Institute for Diseases in the Elderly (RIDE). This study was supported by The Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO) Project No. 050-060-810. This collaborative effort was supported by an award from the National Heart, Lung and Blood Institute (R01-HL-103612, PI BMP).
NEO Study: The authors of the NEO study thank all individuals who participated in the Netherlands Epidemiology in Obesity study, all participating general practitioners for inviting eligible participants and all research nurses for collection of the data. We thank the NEO study group, Pat van Beelen, Petra Noordijk and Ingeborg de Jonge for the coordination, lab and data management of the NEO study. The genotyping in the NEO study was supported by the Centre National de Génotypage (Paris, France), headed by Jean-Francois Deleuze. The NEO study is supported by the participating Departments, the Division and the Board of Directors of the Leiden University Medical Center, and by the Leiden University, Research Profile Area Vascular and Regenerative Medicine. Dennis Mook-Kanamori is supported by Dutch Science Organization (ZonMW-VENI Grant 916.14.023).
Heart and Vascular Health Studies: The Heart and Vascular Health Studies has been funded in part by NHLBI grants R01HL085251 and R01HL073410.
Atherosclerosis Risk in Communities Study: The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
HyperGEN: Genetics of Left Ventricular Hypertrophy: The HyperGEN: Genetics of Left Ventricular Hypertrophy is ancillary to the Family Blood Pressure Program, http://clinicaltrials.gov/ct/show/NCT00005267. Funding sources included National Heart, Lung, and Blood Institute grant R01HL055673 and cooperative agreements (U01) with the National Heart, Lung, and Blood Institute: U01HL054471, U01HL54515 (UT); U01HL054472, U01HL054496 (MN); U01HL054473 (DCC); U01HL054495 (AL); U01HL054509 (NC).
AGES: Age, Gene, Environment, Susceptibility—Reykjavik Study: This study has been funded by NIH contracts N01-AG-1-2100 and 271201200022C, the NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association), and the Althingi (the Icelandic Parliament). The study is approved by the Icelandic National Bioethics Committee, VSN: 00-063. The researchers are indebted to the participants for their willingness to participate in the study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
All studies were approved by local ethics committees and all participants provided written informed consent. The ethics committees for the individual studies are: AGES: The National Bioethics Committee, Iceland; ARIC: University of North Carolina at Chapel Hill Office of Human Research Ethics; CHS: University of Washington Human Subjects Division IRB; HVH: Group Health Cooperative Human Subjects Review Committee; JHS: The Institutional Review Board of the University of Mississippi Medical Center; Medical Center Rotterdam Study: Medical Ethics Committee of the Erasmus Medical Center; HyperGEN: University of Alabama at Birmingham Office of Human Research Ethics; NEO: Ethical committee of the Leiden University Medical Center (LUMC).
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Irvin, M.R., Sitlani, C.M., Noordam, R. et al. Genome-wide meta-analysis of SNP-by9-ACEI/ARB and SNP-by-thiazide diuretic and effect on serum potassium in cohorts of European and African ancestry. Pharmacogenomics J 19, 97–108 (2019). https://doi.org/10.1038/s41397-018-0021-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41397-018-0021-9
This article is cited by
-
Objectives, design and main findings until 2020 from the Rotterdam Study
European Journal of Epidemiology (2020)
-
Genetic correlations and genome-wide associations of cortical structure in general population samples of 22,824 adults
Nature Communications (2020)