Abstract
Background
Hospitalized children with central venous lines (CVLs) are at higher risk of hospital-acquired infections. Information in electronic health records (EHRs) can be employed in training deep learning models to predict the onset of these infections. We incorporated clinical notes in addition to structured EHR data to predict serious bloodstream infections, defined as positive blood culture followed by at least 4 days of new antimicrobial agent administration, among hospitalized children with CVLs.
Methods
Structured EHR information and clinical notes were extracted for a retrospective cohort including all hospitalized patients with CVLs at a single tertiary care pediatric health system from 2013 to 2018. Deep learning models were trained to determine the added benefit of incorporating the information embedded in clinical notes in predicting serious bloodstream infection.
Results
A total of 24,351 patient encounters met inclusion criteria. The best-performing model restricted to structured EHR data had a specificity of 0.951 and positive predictive value (PPV) of 0.056 when the sensitivity was set to 0.85. The addition of contextualized word embeddings improved the specificity to 0.981 and PPV to 0.113.
Conclusions
Integrating clinical notes with structured EHR data improved the prediction of serious bloodstream infections among pediatric patients with CVLs.
Impact
-
Developed an advanced infection prediction model in pediatrics that integrates the structured and unstructured EHRs.
-
Extracted information from clinical notes to do timely prediction in a clinical setting.
-
Developed a deep learning model framework that can be employed in predicting rare events in a complex and dynamic environment.
Similar content being viewed by others
Introduction
Children with central venous lines (CVLs) are at higher risk of the adverse outcomes associated with hospital-acquired infections such as central line-associated bloodstream infection (CLABSI) and sepsis. The U.S. Centers for Disease Control and Prevention estimates that approximately 80,000 new CLABSIs occur in the United States every year, and hospitalized patients who develop CLABSI have a 12–25% increased risk of mortality.1,2
The increasing use of electronic health records (EHRs) in the healthcare domain along with advanced computational techniques lead to the opportunities to create reliable and generalizable population-level monitoring systems that incorporate routinely captured clinical data without the need to conduct resource-intensive chart reviews.3,4,5,6 In recent years, a number of studies have been conducted on the application of advanced analytics of structured EHR data to improve detection and prediction of the adverse outcomes in the hospital.7,8,9 In our own work, we used structured EHR data to predict presumed serious infections (PSIs) and serious bloodstream infections in hospitalized children.4,10
Clinical notes written by health providers are rich sources of a patient’s health status through hospitalization time. While this information has previously been inaccessible to predictive models, more recent natural language processing (NLP) techniques show promise in harnessing the information embedded in unstructured EHRs for aiding clinical decisions.11,12,13 Incorporating structured and unstructured EHR data can boost predictive performance and lead to more accurate results. For example, in adult sepsis prediction, Amrollahi et al. integrated structured and unstructured EHR data to predict the onset of sepsis among intensive care unit (ICU) patients.14 The results showed an improvement in the predictive model’s performance compared to only using the structured EHR data. Similarly, Liang et al. incorporated clinical notes to train a disease classifier to predict a clinical diagnosis for pediatric patients.15 However, these approaches have not been applied to serious bloodstream infections in hospitalized children.
In this study, we investigated the added benefit of integrating structured EHR data with unstructured data gleaned from clinical notes in predicting serious bloodstream infection among hospitalized children with CVLs. We propose a data fusion approach and predictive model that can be employed prospectively in the pediatric ward to predict the risk of a serious bloodstream infection developing during the next 48 h of the hospitalization.
Materials and methods
Study population
Electronic health records, including structured and unstructured data, were extracted for a retrospective cohort of all hospitalized patients with a CVL at a single tertiary care pediatric health system. The inclusion criteria were admission to one of the three freestanding children’s hospitals between January 1, 2013 and December 31, 2018, having a documented CVL at some point during the hospitalization, having length of stay >24 h, and having recorded clinical notes. A complete list of structured information extracted from EHRs is included in Appendix A. This study was approved by the Emory University Institutional Review Board (protocol number 19-012).
Identifying the onset of serious bloodstream infection
PSI was initially proposed as a part of pediatrics sepsis surveillance definition by Hsu et al.16 and has previously been validated.17 Among pediatric patients, PSI was defined as a blood culture drawn and new antibiotic course of at least 4 days (fewer if patients die or are transferred to hospice or another acute care hospital). The minimum of 4 days of antibiotic administration was selected to minimize the false positives from patients for whom the suspected infection was not confirmed and had the empirical treatment stopped. Our primary outcome of serious bloodstream infection was defined as a PSI along with a laboratory confirmed bloodstream infection defined as a positive blood culture.16,18 We reviewed this definition through informal interviews with 2 pediatric infectious disease specialists, 1 pediatric critical care physician, 1 neonatologist, and 1 pediatric hematology/oncology specialist to validate its appropriateness and clinical utility. From this point, we referred to PSI with positive blood culture as serious bloodstream infection (SBI).
Data preprocessing
We used a window-wise study design, as presented in Fig. 1, to predict the onset of SBI in a real-time setting. The start point of the study was the admission time or line insertion time, whichever was earlier. We aimed to predict whether the patient would develop SBI in the next 48-h window; the 48-h prediction window provides enough time for health providers to intervene and potentially prevent a SBI event. In the proposed study design, the SBI prediction was done every 24 h using the most recent information. The 24-h sliding window was selected to ensure that the recorded clinical notes of a patient were updated as the 90th percentile of the time between recording notes for a patient was 23 h. If a 48-h sliding window included an onset of SBI, that window was considered as a positive one. Overall, the prevalence of the positive windows was 0.35% which indicated an extremely imbalanced data problem. Stratified sampling was used to split patient encounters to training (80%) and testing (20%) sets. Moreover, 10% of the training patient encounters were employed as the validation set to optimize the hyperparameters of the models.
If a patient had a documented CVL at the time of admission, the start point of the analysis would be the admission time. Otherwise, the start point would be the first line insertion time. The prediction window was 48 h with a 24 h sliding window until the end of the patient’s hospitalization or removal of the last CVL. When the onset of SBI occurred within a 48 h prediction window, that window was considered positive (red), while the rest (blue) were labeled as negative. The prediction was performed at the start of each arrow.
Structured EHR data
Initially, the structured data included 252 features. The numerical features were transformed, imputed and standardized. The categorical features were one-hot-encoded. We removed multicollinearity with a threshold of 0.8. Finally, there were 129 features from the structured data to include in the analysis. Appendix A includes more details on the preprocessing steps along with a list of the selected features.
Unstructured EHR data
All the provider notes recorded for a patient during the same time-window were concatenated. To reduce the effect of the redundant parts of the clinical notes, we selected the sections with more discriminative information such as history of present illness, impression and plan, patient active problem list, medical decision making, etc. After that, common text preprocessing steps were applied in which all text was transformed to lower case and extra white spaces, punctuations and numbers were removed. Finally, the clinical notes were matched with the corresponding structured data through the text recording timestamps.
Feature extraction from clinical notes
There are two main approaches to incorporate a pre-trained language model in the predictive models; first, fine-tuning a pre-trained language model for down-stream tasks, second, calculating the contextualized word embeddings and feeding them as features to a classification or regression model. We followed the latter approach as it empowered the integration of structured data and clinical notes.
The BERT model has yielded remarkable performance in the clinical domain compared to ELMo and non-contextual embeddings.19 Recent studies have demonstrated that using a domain-specific model achieves better performance compared to nonspecific embeddings; therefore, we employed the Clinical BERT model, which was pre-trained on approximately two million clinical notes in MIMIC-III dataset,20 to acquire the contextualized word embeddings for the clinical notes in our cohort.12 To assess the performance of the contextual word embeddings from the Clinical BERT model, we also extracted text features through the term frequency-inverse document frequency (TF-IDF) method.
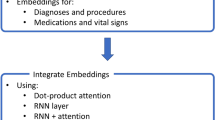
Figure 2 demonstrates our approach to integrate the clinical notes with the rest of the structured clinical features to train the predictive models.
The most informative sections of the clinical notes recorded for a patient at the time of prediction were selected and provided to the Clinical BERT model to calculate the contextualized word embeddings using the last hidden layer of the model. Then the 768-dimensional contextualized word embeddings were concatenated with the 129-dimensional features from the structured EHR at every prediction point. The Bidirectional LSTM model with the attention mechanism and Focal loss incorporated the 897-dimensional input to predict if a SBI will occur during the next 48 h of this patient’s hospitalization.
Predictive models
Model structure
We employed Bidirectional Long Short-Term Memory (BiLSTM) model as BiLSTMs can look at the information prior and successor of a given word in the note, which is closer to human reading abilities and yields strong performance in the NLP domain.14
Loss function
There are two types of observations in a classification task: hard and easy. The hard observations are defined as the ones that confuse the predictive model. These are the examples that the model should focus on to improve its overall performance. The extreme class-imbalanced problem in this study (prevalence of 0.35%) required a strategy to assign more weight to the minority class observations while taking the easy/hard examples into consideration to ultimately improve the true positive and true negative predictions. We employed Focal loss as a solution to this obstacle.21 Focal loss is a loss function to lessen the weight of easy examples while intensifying the penalization in the case of an incorrect classification of hard examples.
Attention mechanism
Attention mechanism in deep learning was motivated by how humans pay attention to different regions of an image or correlate words in a sentence.22 When it comes to a class-imbalanced classification task, it is crucial to attend to the more prominent parts of the input sequence to achieve better results. Since we had long sequences of structured and unstructured data, the attention mechanism was incorporated to train the model to further attend to the more relevant parts of the input and explain the relationship between words in the context.
Training process
We trained five models with the following input features to evaluate the benefit of integrating structured EHRs with clinical notes; (1) structured data, (2) extracted features from unstructured data with TF-IDF, (3) extracted contextualized word embeddings from unstructured data using Clinical BERT, (4) structured data along with the TF-IDF features, (5) structured data and the contextualized word embeddings. We trained all the models with a batch size of 128, Adam optimizer, and dropout regularization. The hyperparameters of the models (e.g., learning rate, dropout rate, number of neurons for each layer, etc.) were tuned by Bayesian optimization method. Appendix B includes the details on model training, structure, and optimization.
Statistical analysis
To check the statistically significant difference of features’ values between the SBI and non-SBI groups, Wilcoxon rank-sum test for numerical features and Chi-squared test for categorical features were applied. Moreover, the estimated 95% confidence interval (CI) of the models’ performance metrics was calculated through bootstrapping method.
This manuscript was prepared using the guidelines provided by Leisman et al.23 for reporting of prediction models.
Results
For this study, 97,424 patient encounters associated with 15,704 patients were extracted from the EHRs. Among these patient encounters, there were outpatient appointments and hospital outpatient department visits for patients with existing CVLs; therefore, 73,073 patients encounters were excluded from the cohort due to length of stay <24 h or not having recorded clinical notes. After applying these exclusion criteria, a total number of 2733 neonates (age <28 days), 5383 infants (age between 28 days and 1 year), 4286 toddlers and preschoolers (age between 1 and 5 years), 5625 children (age between 5 and 12 years), and 6324 adolescents (age >12 years) were included in the analysis. Figure 3 demonstrates the associated CONSORT diagram.
The demographic and clinical characteristics of the patients in our study are listed in Table 1. According to the results, SBI patients were younger (median = 3.1 vs. 5.8 years, p < 0.001), with lower weights (13.6 vs. 19.3 Kg, p < 0.001), shorter heights (90.3 vs. 110.2 cm, p < 0.001), more African Americans (44.3 vs. 36.3%, p < 0.001), and less Caucasian (47.1 vs. 54.1%, p = 0.002). Overall, SBI patients had higher hospital length of stay (36.7 vs. 6.1 days, p < 0.001), had higher ICU admissions (65.4 vs. 48.2%, p < 0.001), and had a higher rate of having Medicaid health insurance (62.5 vs. 57.1%, p = 0.02) while having a lower rate in Commercial health insurance (34.2 vs. 38.9%, p = 0.03). The mortality rate was higher among SBI patients but there were no statistically significant differences for this feature among the two groups (0.2 vs. 0.06%, p = 0.22). Other features were comparable between SBI and non-SBI groups (p > 0.05).
Statistical tests were performed to assess statistical significance between the components of PEdiatric Logistic Organ Dysfunction (PELOD-2) score and Pediatric RISk of Mortality (PRISM-III) Score between SBI and non-SBI groups across time windows.24,25 PELOD-2 was primarily designed to describe the severity of organ dysfunction and PRISM-III was developed to predict the risk of mortality among the pediatric population. The results are presented in Appendix D and E.
To assess the benefit of incorporating the clinical notes in SBI prediction, five predictive models with different inputs were trained. The performance metrics are presented in Table 2. According to these results, the model which coupled the structured clinical features with word embeddings from Clinical BERT model outperformed the rest of the models with highest specificity of 0.981 with 95% CI = [0.980, 0.982], positive predictive value (PPV) of 0.113 [0.09, 0.137], negative predictive value (NPV) of 0.999 [0.999, 0.999], accuracy of 0.980 [0.978, 0.981], F-1 score of 0.195 [0.159, 0.231], and area under the precision–recall curve (AUPRC) of 0.282 [0.188, 0.366]. Figures 4 and 5 demonstrate the associated receiver operative characteristics and precision–recall curves for all the five models applied on the testing dataset, respectively.
Using the word representation from the last four hidden layers of the Clinical BERT model (instead of only using the last hidden layer) did not improve the models’ performance while it added to the computational costs (Appendix C).
Discussion
In this study, we evaluated the effect of coupling clinical notes with the structured clinical features (e.g., demographic, physiological, laboratory test results, etc.) in predicting the onset of SBI, defined as a culture drawn associated with a positive test result followed by at least 4 days of new antimicrobial agent administration, among pediatric patients with CVLs. The proposed deep learning model predicts if a hospitalized patient with CVL will develop SBI during the next 48 h of hospitalization. Our model had a PPV of 0.113, which is 32 times greater the baseline prevalence of SBI across the 48-h time windows, and a very high NPV 0.999, which presents the strength of the model in ruling out the patients with lower risk of the infection. Incorporating the clinical notes improved the specificity (0.951 vs. 0.981, p value <0.001), PPV (0.056 vs. 0.113, p value <0.001), accuracy (0.950 vs. 0.980, p value <0.001), F-1 score (0.105 vs. 0.195, p value <0.001), and AUPRC (0.202 vs. 0.282, p value <0.001) compared to the model that employed only the information from structured EHRs.
The predictive performance of our proposed model and study design outperformed the performance of the prior models trained to predict CLABSI. Most of these models were based on a retrospective case control study and did not incorporate the temporal information in EHRs. Training a Random Forest predictive model based on non-temporal data, Beeler et al. obtained AUROC of 0.87 in predicting CLABSI among adult, pediatric, and neonatal patients.26 Sung et al. trained a CLABSI prediction model using Gradient Boosting Trees which attained AUROC of 0.77 among pediatric cohorts receiving cancer medications.27 Our model and study design had characteristics that may have contributed to achieving better predictive performance; first, we only included the pediatric patients with a documented CVL at the time of admission or at some point during hospitalization. Second, we extracted and incorporated an extensive set of features recorded in EHR. Third, we used the information embedded in the clinical notes through a state-of-the-art NLP framework. Finally, we trained a deep learning model capable of including the temporal information while dealing with low prevalence classification problem by using a loss function specifically designed for extreme class-imbalanced classification and attention mechanism to focus on the most predictive parts of the input sequence at every prediction point.
Previous studies have been done to investigate the added benefit of integrating clinical notes to the structured EHR data in predicting patient clinical outcomes. Amrollahi et al. utilized structured and unstructured EHR information to model capable of timely prediction of sepsis which outperformed the model trained only on the structured data.14 In a similar study, Goh et al. developed an artificial intelligence algorithm incorporating the two data modalities and concluded that the model performance improved after integrating the clinical notes to the input features.28 In another study, Horng et al. conducted a research to demonstrate the incremental benefit of using free text data in addition to vital sign and demographic data to identify patients with suspected infection in the emergency department.29
Limitations
Our study has several limitations. First, while we intentionally extracted EHR features that are routinely recorded across systems, the external application of the proposed model, which was trained within a single pediatric health system, on other health systems may be biased. Second, we utilized two structured EHR variables, the timestamp associated with antibiotics administration and positive blood culture specimen taken time, to identify the onset of SBI among this cohort; therefore, our outcome definition led to refining the cohort by structured data. Third, the deep learning field is very dynamic and new models are introduced every day; therefore, the model structure that we applied to tackle low prevalence classification problems and extract contextualized word embeddings may not reflect all the capacity of deep learning application in predicting this adverse outcome. Fourth, in some cases, the clinical notes are updated with delay. This delay in recording clinical notes may affect the performance of the model. Finally, the Clinical BERT model used for extracting contextualized word embeddings has a limitation in the number of words it can take in each of the recorded clinical notes at every prediction point. This limitation requires extensive text preprocessing to only include the parts describing the patient’s health condition at the moment and exclude history and administrative sections.
Conclusion
In this study, we demonstrated the potential of information embedded in clinical notes in predicting SBI among pediatric patients with CVLs through a deep learning approach. The results indicated the superior performance of the model that integrated the structured and unstructured EHRs and the possible benefit in predicting patient outcome in a clinical setting.
Data availability
The dataset analyzed during the current study were extracted from a healthcare system and contained patient-protected health information; therefore, the dataset cannot be publicly shared.
References
Rupp, M. E. & Majorant, D. Prevention of vascular catheter-related bloodstream infections. Infect. Dis. Clin. 30, 853–868 (2016).
Centers for Disease Control and Prevention. Vital signs: central line–associated blood stream infections—United States, 2001, 2008, and 2009. Ann. Emerg. Med. 58, 447–450 (2011).
Walker, L. W., Nowalk, A. J. & Visweswaran, S. Predicting outcomes in central venous catheter salvage in pediatric central line–associated bloodstream infection. J. Am. Med. Inf. Assoc. 28, 862–867 (2021).
Tabaie, A. et al. Predicting presumed serious infection among hospitalized children on central venous lines with machine learning. Comput. Biol. Med. 132, 104289 (2021).
Le, S. et al. Pediatric severe sepsis prediction using machine learning. Front. Pediatr. 7, 413 (2019).
Masino, A. J. et al. Machine learning models for early sepsis recognition in the neonatal intensive care unit using readily available electronic health record data. PLoS ONE 14, e0212665 (2019).
Reyna, M. A. et al. Early prediction of sepsis from clinical data: the PhysioNet/Computing in Cardiology Challenge 2019. In 2019 Computing in Cardiology (CinC). pp. 1 (IEEE, 2019).
Nemati, S. et al. An interpretable machine learning model for accurate prediction of sepsis in the ICU. Crit. Care Med. 46, 547 (2018).
Raita, Y. et al. Machine learning-based prediction of acute severity in infants hospitalized for bronchiolitis: a multicenter prospective study. Sci. Rep. 10, 10979 (2020).
Tabaie, A. et al. Deep learning model to predict serious infection among children with central venous lines. Front. Pediatr. 9, 726870 (2021).
Huang, K., Altosaar, J. & Ranganath, R. ClinicalBERT: modeling clinical notes and predicting hospital readmission. Preprint at arXiv https://arxiv.org/abs/1904.05342 (2019).
Alsentzer, E. et al. Publicly available clinical BERT embeddings. Preprint at arXiv https://arxiv.org/abs/1904.03323 (2019).
Zhang, D., Thadajarassiri, J., Sen, C. & Rundensteiner, E. Time-aware transformer-based network for clinical notes series prediction. In Machine Learning for Healthcare Conference 566–588 (PMLR, 2020).
Amrollahi, F., Shashikumar, S. P., Razmi, F. & Nemati, S. Contextual embeddings from clinical notes improves prediction of sepsis. AMIA Annu. Symp. Proc. 2020, 197–202 (2020).
Liang, H. et al. Evaluation and accurate diagnoses of pediatric diseases using artificial intelligence. Nat. Med. 25, 433–438 (2019).
Hsu, H. E. et al. A national approach to pediatric sepsis surveillance. Pediatrics 144, e20191790 (2019).
Weiss, S. L. et al. Identification of pediatric sepsis for epidemiologic surveillance using electronic clinical data. Pediatr. Crit. Care Med. 21, 113–121 (2020).
Rhee, C., Dantes, R. B., Epstein, L. & Klompas, M. Using objective clinical data to track progress on preventing and treating sepsis: CDC’s new ‘Adult Sepsis Event’ surveillance strategy. BMJ Qual. Saf. 28, 305–309 (2019).
Si, Y., Wang, J., Xu, H. & Roberts, K. Enhancing clinical concept extraction with contextual embeddings. J. Am. Med. Inf. Assoc. 26, 1297–1304 (2019).
Johnson, A. E. et al. MIMIC-III, a freely accessible critical care database. Sci. Data 3, 1–9 (2016).
Lin, T. Y., Goyal, P., Girshick, R., He, K. & Dollár, P. Focal loss for dense object detection. In Proc. IEEE International Conference on Computer Vision 2980–2988 (IEEE, 2017).
Bahdanau, D., Cho, K. & Bengio, Y. Neural machine translation by jointly learning to align and translate. Preprint at arXiv https://arxiv.org/abs/1409.0473 (2014).
Leisman, D. E. et al. Development and reporting of prediction models: guidance for authors from editors of respiratory, sleep, and critical care journals. Crit. Care Med. 48, 623 (2020).
Leteurtre, S. et al. PELOD-2: an update of the PEdiatric logistic organ dysfunction score. Crit. Care Med. 41, 1761–1773 (2013).
Pollack, M. M., Patel, K. M. & Ruttimann, U. E. PRISM III: an updated Pediatric Risk of Mortality score. Crit. Care Med. 24, 743–752 (1996).
Beeler, C. et al. Assessing patient risk of central line-associated bacteremia via machine learning. Am. J. Infect. Control 46, 986–991 (2018).
Sung, L. et al. Development and utility assessment of a machine learning bloodstream infection classifier in pediatric patients receiving cancer treatments. BMC Cancer 20, 1–9 (2020).
Goh, K. H. et al. Artificial intelligence in sepsis early prediction and diagnosis using unstructured data in healthcare. Nat. Commun. 12, 711 (2021).
Horng, S. et al. Creating an automated trigger for sepsis clinical decision support at emergency department triage using machine learning. PLoS ONE 12, e0174708 (2017).
Funding
This project was supported by an institutional grant provided by the Children’s Healthcare of Atlanta, Atlanta, GA through The Pediatric Technology Center, in conjunction with the Health Analytics Council. All views expressed in this article are the authors’ own and do not necessarily reflect the views of the authors’ employers and funding bodies.
Author information
Authors and Affiliations
Contributions
Substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data: A.T., E.W.O., R.K. Drafting the article or revising it critically for important intellectual content: A.T., E.W.O., S.K., R.K. Final approval of the version to be published: A.T., E.W.O., S.K., R.K.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
A waiver of informed consent was requested and approved by the Institutional Review Board.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Tabaie, A., Orenstein, E.W., Kandaswamy, S. et al. Integrating structured and unstructured data for timely prediction of bloodstream infection among children. Pediatr Res 93, 969–975 (2023). https://doi.org/10.1038/s41390-022-02116-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-022-02116-6