Abstract
Background
Preterm infants are generally fed through nasogastric enteral feeding tubes (NEFTs). The aim of this work was to evaluate the role of NEFTs in the initial colonization of the preterm gut and its evolution within the first 2 weeks after birth.
Methods
For this purpose, fecal and NEFT-derived samples from 30 preterm infants hospitalized in a neonatal intensive care unit (NICU) were collected from birth to the second week of life. Samples were cultivated in ten culture media, including three for the isolation of antibiotic-resistant microorganisms.
Results
Isolates (561) were identified by 16S ribosomal RNA gene sequencing. Although the first NEFTs inserted into the neonates after birth were rarely colonized, analysis of NEFTs and fecal samples over time revealed a significant increase in bacterial abundance, diversity, and detection frequency. Results showed a parallel colonization between time-matched NEFTs and fecal samples, suggesting an ongoing bidirectional transfer of bacteria from the neonatal gut to the NEFTs and vice versa.
Conclusions
In short-term hospitalization, length is by far the determinant factor for the early colonization of preterm infants. As NEFT populations reflect the bacterial populations that are colonizing the preterm in a precise moment, their knowledge could be useful to prevent the dissemination of antibiotic-resistant strains.
Impact
-
The hospital environment modulates preterm colonization immediately after birth.
-
The colonization of preterm feces and NEFTs occurs in parallel.
-
There is an ongoing bidirectional transfer of microorganisms from the neonatal gut to the NEFTs and vice versa.
-
Bacterial communities inside NEFTs could act as reservoirs of antibiotic resistance genes.
-
NEFT populations reflect the bacteria that are colonizing the preterm at a precise moment.
Similar content being viewed by others
Introduction
Prematurity is the main cause of mortality in children under five years.1 Preterm infants are exposed to a stressful vital experience early in life, including maternal separation, frequent medical interventions, and a harsh environment, in which nosocomial microorganisms may represent a serious health threat.2,3 Overall, these factors have a profound impact on the establishment of their intestinal microbiota.4,5,6,7,8
The suck-swallow mechanism is not fully developed at birth in many preterm neonates.9 Consequently, their nutrition may be dependent on nasogastric enteral feeding tubes (NEFTs) for some weeks. Despite following good hygienic practices, a plethora of microorganisms unavoidably and rapidly colonize the surfaces of NEFTs and other invasive devices. The duration of a single NEFT placement depends on the institution and it can be shortened if necessary in case of suspicion of an infection and/or tube clogging. Previous work has demonstrated that microorganisms usually require <48 h to colonize their inner surface.10
The NEFT-associated microbiota reflects the environment of each patient and includes both vertically and horizontally transmitted microorganisms.11,12,13 Concretely, preterm-associated NEFTs are exposed to the oral, nasopharyngeal, and skin microbiota of the infant, to the microorganisms carried by the food they receive (own mother’s milk [OMM], donor human milk [DHM], and/or infant formula), to those harbored by the mother and other relatives, the medical staff and their clothing, and to those present in aerosols and surfaces.14,15 Imaging of the internal surfaces of NEFTs has revealed that the microbes present in such devices are forming complex biofilms.16,17 Once such structures are formed, microorganisms may become highly resistant to antimicrobial agents, including antibiotics, antiseptics, and disinfectants,18 and, consequently, they can act as a shelter for high-risk nosocomial clones within the neonatal intensive care units (NICUs).19,20,21 Dispersed cells and clumps detached from the biofilm can be introduced into the preterm gastrointestinal tract every time that feeding is injected through the NEFT. This situation may increase the dysbiosis state that usually characterizes the preterm gut microbiota and, ultimately, lead to infections and sepsis.11,22 Indeed, many infectious diseases in preterm infants have been linked to contamination of NEFTs and other medical devices.23,24
On the other hand, some authors have suggested that NEFTs colonization is partly due to gastroesophageal reflux, which is physiological in newborns because of cardiac immaturity.25 This can be aggravated by both the placement of an NEFT that keeps the lower esophageal sphincter continuously open and the supine position of the preterm infant.26 Indeed, the same bacterial strains have been found both in milk (after passing through NEFTs) and in the feces of preterm infants.16 Similarly, microorganisms from the oropharynx of elderly patients are particularly frequent in the gastric juice of those fed through an NEFT, suggesting that these devices may act as the connectors for a bidirectional transmission between these two anatomic sites.27
Several studies have evaluated the type of microorganisms related to preterm-inserted NEFTs depending on different factors, such as the type of feeding.25,28,29,30 However, the role of the first NEFT inserted into a neonate immediately after birth in the early gut colonization remains unknown. In this context, the main objective of this study was to evaluate the initial bacterial colonization of the first NEFT in hospitalized preterm infants and the evolution of the bacterial populations in subsequent NEFTs and in their feces using a culturomic approach. The impact of the type of feeding, the antibiotherapy, and the use of probiotics in the colonization pattern of NEFT-derived and fecal samples was also evaluated.
Materials and methods
Ethics statement
The Ethical Committee on Clinical Research of La Paz University Hospital, Madrid (Spain) approved the study (Code: HULP PI-3199; date 27 June 2018). The collection of samples and clinical information was done after gathering the written consent from the legal guardians of the children who participated in this study, who were previously informed of all the protocols.
Subjects and study design
This study included 30 preterm children born at the La Paz University Hospital, Madrid (Spain) from April to October 2018, whose demographic characteristics and relevant clinical data are summarized in Table 1. Subjects recruited for this study were born at a gestational age <32 weeks and admitted to the NICU. Exclusion criteria included preterm infants with malformations or congenital metabolic diseases. Enteral feeding was introduced after birth using NEFTs. All participants were fed by gravity, every 3 h and eight times a day (as a general rule). More details regarding nutrition policy in the NICU are included in the Supporting information file (S1). Infants were arbitrarily classified into three groups according to the type and the proportion of feeding received, as follows (Table 1): (a) those for whom OMM represented >75% of the feeds (OMM group; n = 13); (b) those for whom DHM represented >75% of the feeds (DHM group; n = 5); and (c) those receiving OMM and/or DHM in other proportions and/or formula milk (mixed feed, MF group; n = 12). Feeding type was daily assessed all over the study. The feeding of 12 infants (40%) was supplemented with Limosilactobacillus reuteri (formely known as Lactobacillus reuteri) DSM 17938 (BioGaia ProTectis drops) (five drops a day, equivalent to 108 CFU).
Samples of meconium (Me), feces, and NEFTs were collected over the first 17 days of life of the preterm following the experimental design shown in Fig. 1. The first spontaneously evacuated Me was collected at day 1.1 (0.3–1.8) (median [interquartile range [IQR]), the first set of samples (NEFT-1 and F1) were collected on day 3. The second set of samples (NEFT-2 and F2) were collected on day 9 and the third set (NEFT-3 and F3) on day 17 (see Supplementary Table S1 for individual data). All NEFTs obtained in this study were inserted into the infants (per protocol) for 2.0 (2.0–2.2) days (median [IQR]) (Fig. 1; see Supplementary Table S2 for individual data).
Fecal (F) and nasogastric enteral feeding tube (NEFT) samples were collected over the first 17 days of life of 30 preterm children. Days highlighted in light grey correspond to those of NEFTs insertion. The median (IQR) time (days) of this step is included in the boxes below. Days highlighted in light orange correspond to those in which samples (F and NEFTs) were taken. Median (IQR) values of the time at sample collection (days) are included in the boxes marked with the same color. In grey is represented the day samples of the first spontaneously evacuated meconium (Me) were taken. In the box below is included the median (IQR) value of the time (days) at Me collection. In all the children, NEFT-1 is the first device inserted into the preterm after birth. In the end, for every participant, the following set of samples were collected: one sample of Me and three sets of NEFT-F matching samples at day 3 (NEFT-1 and F1), day 9 (NEFT-2 and F2), and day 17 (NEFT-3 and F3) of the study.
All samples were collected by the medical staff and nurses of the Neonatology Unit of the hospital. Fecal samples were collected from the diaper of each infant using disposable sterile spatulas, placed in a sterile tube, and stored at −80 °C until processing. NEFTs were removed from the infants at different times (Fig. 1) after the last feeding received (3 h post-feeding as standard, except in case of hypoglycemia, in which feeding needs to be more frequent and this interval could have been shortened) using sterile gloves, placed into sterile plastic bags and frozen at −80 °C. Collected samples were then transferred to the laboratory where they were kept at −80 °C until processing. An ID number was assigned to all the NEFTs and fecal samples to unequivocally carry out data analysis at the end of the study.
Sample processing
For bacterial analysis of NEFTs, the external part of NEFTs was first cleaned with 70% (v/v) ethanol. To determine the bacterial population inside the NEFTs, a three-step procedure was followed. The first two steps were based on the protocol described by Petersen et al.10 In the first step, the residual liquid inside the tube lumen was flushed into a 50 ml tube. Then, 4 ml of 0.85% (w/v) sterile saline were pumped into the NEFT with a syringe to remove loosely attached microorganisms, and the collected fluid was added to the first tube. Finally, we proceed to remove the biofilm from the inner surface of the NEFT; for this purpose, the cleaned device was first cut into several pieces of ~2 cm length using sterilized scissors. Then, all the pieces were placed into a tube containing 10 ml of sterile saline and the tube was vortexed for 1 min. The suspension was mixed with the fluid obtained after the first two steps and centrifuged at 31,231 × g for 10 min at 4 °C. The pellet was suspended in 4 ml of sterile saline and divided into four aliquots (1 ml each), which were stored at −80 °C for further analysis.
Culture analysis and identification of the isolates
To determine the viable bacterial counts in NEFTs, the cell suspension obtained in the section “Sample processing,” and immediately after processing (i.e., prior freezing) was decimally diluted, and 20 μl of the pertinent dilutions were plated on different culture media: Man, Rogosa, and Sharpe (MRS; Oxoid, Basingstoke, UK) agar plates supplemented with l-cysteine (0.5 g/l) (MRScys) (Sigma, St. Louis, MO) for isolation of lactobacilli and other lactic acid bacteria; Columbia agar with nalidixic acid and colistin (CNA, Oxoid) for isolation of Gram-positive cocci; Pseudomonas Agar Base (PAB, Oxoid) for isolation of Pseudomonas spp.; and MacConkey (MCK, Oxoid) for isolation of Enterobacteriaceae. In addition, Brilliance MRSA 2 Agar (MRSA), Brilliance VRE Agar (VRE), and Brilliance CRE Agar (CRE) (Oxoid) were also included for the isolation of methicillin-, vancomycin-, and carbapenem-resistant bacteria, respectively. All plates were incubated for 48 h at 37 °C under aerobic conditions, except MRScys plates that were incubated anaerobically (85% nitrogen, 10% hydrogen, and 5% carbon dioxide) in an anaerobic workstation (MINI-MACS, DW Scientific, Shipley, UK). For microbial analysis of infant feces, 0.1 g of each stool sample was mixed with 0.9 g of peptone water. Once fully homogenized, the sample was decimally diluted and 20 μl of the pertinent dilutions were spread on the culture media described above. Bacterial counts were expressed as colony-forming units (CFU) per ml of Me or feces, or and as CFU/cm2 in the case of the NEFT content.
At least one representative of each colony morphology was selected from the agar plates and spread on Brain Heart Infusion (Oxoid) or MRScys agar plates to check for purity. Identification of the isolates at the species level was carried out by Sanger sequencing of the 16S ribosomal RNA (rRNA) gene following the protocol described in ref. 31. For this, purified PCR products were sequenced (STAB VIDA, Caparica, Portugal) and after obtaining the partial 16S rRNA region nucleotide sequence, all sequences were identified using BlastN (version 2.10) with the 16S rRNA database of NCBI.
Statistical analysis
Microbiological data (recorded as CFU/ml) were transformed to logarithmic values before statistical analysis. The statistical analyses were performed with the R software (×64) 4.0.3 desktop version. The normality of data distribution was analyzed using the Shapiro–Wilk test, evidencing non-normal distribution for all tested variables (p < 0.05). Accordingly, concentrations of microorganisms were expressed as median and IQR values and analyzed using nonparametric statistical tests. Differences over time in median bacterial counts in NEFTs and fecal samples where bacterial growth was detected in the culture media tested were evaluated using Friedman’s tests with pairwise comparisons using Nemenyi post hoc tests for unreplicated blocked data using the R package PMCMRplus.32 Detection frequency (n) comparisons over time in NEFTs and fecal samples in the culture media tested were performed using the Cochran’s Q tests with false discovery rate post hoc tests for pairwise comparisons using the R packages RVAideMemoire33 and rcompanion,34 respectively.
In order to evaluate differences in the microbial populations in NEFTs and feces in the culture media tested according to the type of feeding, the antibiotic therapy, and the probiotic treatment, the Wilcoxon’s rank tests for variables with two factors and the Kruskal–Wallis tests for variables with three factors were applied using Bonferroni p value adjustment for pairwise comparisons. Differences were considered significant at p < 0.05 for all analyses. Heatmaps were performed with the Gplots package 3.1.1 version of the R software.35 The cladograms were performed with the Hclust hierarchical cluster analysis with complete linkage method from the R’s core package “stats”. To estimate the diversity of the samples, Shannon and Simpson diversity indexes were performed with the R vegan package (version: 2.5.6).36 Comparisons over time of diversity indexes in NEFT and fecal samples were performed with the Friedman’s test with pairwise comparisons using Nemenyi post hoc tests.
Results
Culture analysis of NEFTs and fecal samples
In total, 90 NEFT-derived and 120 Me/fecal samples were collected. In Fig. 2 are displayed the median (IQR) counts over time on different culture media in NEFTs (a) and in Me/fecal (b) samples where growth was detected.
Comparisons over time of median counts on different culture media in NEFTs (a) and fecal (b) samples where bacterial growth was detected. Boxes show the median and interquartile range (IQR) of the samples where growth was detected. Numbers in brackets below each time point indicate the number of samples where bacterial growth was detected (N = 30) (see Supplementary Tables S4 and S5 for statistical differences over time assessed by Cochran’s Q test with post hoc FDR adjustment for pairwise comparisons). P values show statistically significant differences in median values along time (*p < 0.05; ***p < 0.001) assessed by Friedman’s test (see Supplementary Tables S4 and S5 for pairwise comparisons of median values with Nemenyi’s adjustment). CNA: Columbia agar with nalidixic acid and colistin for isolation of Gram-positive cocci, MRScys: Man, Rogosa, and Sharpe agar plates supplemented with l-cysteine (0.5 g/l) for isolation of lactobacilli and other lactic acid bacteria, MRSA Brilliance: MRSA 2 Agar for the isolation of methicillin-resistant bacteria, VRE Brilliance: VRE Agar for the isolation of vancomycin-resistant bacteria, MCK: MacConkey for isolation of Enterobacteriaceae; PAB: Pseudomonas Agar Base for isolation of Pseudomonas spp., CRE Brilliance: CRE Agar for isolation of carbapenem-resistant bacteria.
Regarding NEFT-1 samples (N = 30) bacterial populations were dominated by Gram-positive microorganisms detected in 33%, 13%, and 23% of the samples plated on CNA, MRScys, and MRSA, respectively, with a median count of about 3 log10 CFU/cm2. Growth was not detected in four of the culture media tested: VRE, MCK, PAB, and CRE (Fig. 2a). The detection frequency (n) or the number of samples where bacterial growth was detected significantly increased from sampling times 1 to 2 (p ≤ 0.001), except in VRE and CRE plates where bacterial growth was only detected in one and seven NEFTs samples, respectively (Fig. 2a and Supplementary Table S4). Bacterial median counts followed the same trend, increasing significantly (p < 0.001) from NEFT-1 to NEFT-2, particularly the populations of Enterobacteriaceae, Pseudomonas spp., and carbapenem-resistant microorganisms (Fig. 2a and Supplementary Table S4).
As for fecal samples, vancomycin- and carbapenem-resistant microorganism were not recovered from Me and F1 samples, respectively. In all the culture media, fecal median counts increased significantly over time (p < 0.001), especially from sampling times 1 to 2 (Fig. 2b and Supplementary Table S5). At the end of the study (F3) median counts in the three media for the detection of resistant microorganisms (i.e., MRSA, VRE, and CRE) were 5.6, 4.3, and 4.0 log10 CFU/ml, respectively (Fig. 2b and Supplementary Table S5).
A heatmap of the NEFT samples according to the bacterial growth in the different media showed four well-defined groups (Fig. 3). The central cluster (cluster I) included most of the collected NEFT-1 samples, which was characterized by the absence of cultivable bacteria in the media and incubation conditions tested in this study. A second cluster (cluster II) included samples belonging to the three sampling times although it was richer in NEFT-1 and NEFT-2 samples. They were characterized by the presence of Gram-positive bacteria (able to grow on CNA) and, less frequently, of methicillin-resistant microorganisms. A third group (cluster III) included most of the NEFT-2 samples and some of the NEFT-1 and NEFT-3 ones, displaying an abundance of Gram-positive microorganisms able to grow on CNA, MRScys, and MRSA plates and the absence or low counts (<2 log10 CFU) in the media supporting growth of Gram-negative bacteria. In contrast, the fourth cluster (cluster IV) included most of the NEFT-3 samples, which showed a wider diversity and higher Enterobacteriaceae (MCK) counts (Fig. 3).
This heatmap shows the relatedness between NEFT samples of 30 preterm infants (N = 90) according to their microbial counts in different culture media. The purple color indicates NEFT-1 samples (i.e., the first NEFT inserted after birth), the pink color indicates NEFT-2 samples (day 9), and the light orange color indicates NEFT-3 samples (day 17). On the right, changes in color indicate microbial counts expressed as log10 CFU. As stated in the key panel, green color means absence, and white color means samples with higher counts. On the right, the fourth clusters have been highlighted. CNA: Columbia agar with nalidixic acid and colistin for isolation of Gram-positive cocci, MRSc: Man, Rogosa, and Sharpe agar plates supplemented with l-cysteine (0.5 g/l) for isolation of lactobacilli and other lactic acid bacteria, MRSA Brilliance: MRSA 2 Agar for the isolation of methicillin-resistant bacteria, VRE Brilliance: VRE Agar for the isolation of vancomycin-resistant bacteria, MCK: MacConkey for isolation of Enterobacteriaceae, PAB: Pseudomonas Agar Base for isolation of Pseudomonas spp., CRE Brilliance: CRE Agar for isolation of carbapenem-resistant bacteria.
Fecal samples were also clustered in four groups (Fig. 4). Cluster I grouped most of the Me and F1 samples. Cluster II showed a profile similar to NEFT-related cluster II (concretely, Gram-positive bacteria able to grow in CNA and MRSA). Cluster III included some F1 and F2 samples, while cluster IV grouped F2 and F3 samples with a profile similar to that of NEFT-related cluster II.
This heatmap shows the association between microbial counts in different culture media and stool samples of 30 preterm infants (N = 120). The blue color indicates samples of meconium (Me), the red color indicates fecal samples at day 3 (F1), the light orange color indicates fecal samples at day 9 (F2), and the green color indicates fecal samples at day 17 (F3). On the right, changes in color indicate microbial counts expressed as log10 CFU. As stated in the color key panel, the green color means absence and the white color means samples with higher counts. On the right, the fourth clusters have been highlighted. CNA: Columbia agar with nalidixic acid and colistin for isolation of Gram-positive cocci, MRSc: Man, Rogosa and Sharpe agar plates supplemented with l-cysteine (0.5 g/l) for isolation of lactobacilli and other lactic acid bacteria, MRSA Brilliance: MRSA 2 Agar for the isolation of methicillin-resistant bacteria, VRE Brilliance: VRE Agar for the isolation of vancomycin-resistant bacteria, MCK: MacConkey for isolation of Enterobacteriaceae, PAB: Pseudomonas Agar Base for isolation of Pseudomonas spp., CRE Brilliance: CRE Agar for isolation of carbapenem-resistant bacteria.
Diversity of bacterial isolates in NEFT and fecal samples
A total of 561 isolates were obtained, including 205 (37%) from NEFTs and 356 (63%) from Me and feces. In relation to NEFTs, 25 (12%), 63 (31%) and 117 (57%) isolates were recovered from NEFT-1, NEFT-2, and NEFT-3 samples, respectively. In relation to fecal samples, 61 (17 %) isolates were obtained from Me and F1 samples, 117 (33%) from F2, and 178 (50%) from F3 (Fig. 5a).
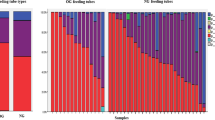
A pie charts (a) showing the number and percentage of isolates in each type of sample. b Distribution of relative abundance of the isolates at the genus level per sample. Each of the stacked bar plots illustrates the relative abundance (y-axis) of the isolated species in the 30 NEFTs and fecal samples over time. Isolates with a relative abundance <1 % were included in the “other“ group. c Detection rate (number of positive samples in which a particular bacterial genus or species was detected at each sampling time) of isolates in the NEFTs and fecal samples over time. d Distribution of the relative abundance of Staphylococcus spp. isolates per sample. Each of the stacked bar plots illustrates the relative abundance (y-axis) of isolated species in the NEFTs and fecal samples over time.
Identification of the isolates revealed that most of them belonged to 11 genera (Fig. 5b). The genus Staphylococcus was the most abundant (216 out of the 561 isolates) in all the samples independently of the collecting time (Fig. 5b), being particularly dominant in Me and F1 samples (74% and 54 % of the isolates, respectively). The relative abundance of the 11 species belonging to this genus is shown in Fig. 5d. Independently of the sampling time, Staphylococcus epidermidis was the most abundant species, followed by Staphylococcus capitis and Staphylococcus haemolyticus. Other staphylococcal species were sporadically isolated as the hospitalization time increased. The profiles of staphylococcal species found in NEFTs samples were similar to those observed in their matched fecal samples (Fig. 5d). Isolates belonging to the genera Enterococcus and Lactobacillus were also identified in these samples, although their detection frequencies were significantly lower (p < 0.05) (Fig. 5c). The presence of members of the genus Enterococcus (75 isolates) and the family Enterobacteriaceae (128 isolates) increased over time. Among the family Enterobacteriaceae, the genus Klebsiella was the most abundant (39 isolates) (Fig. 5c).
Both types of samples showed a significant increase in bacterial diversity from sampling times 1 to 3 (p < 0.05) assessed using the Shannon and Simpson diversity indices (median [MIN–MAX]) (Supplementary Table S6 and Supplementary Fig. S1). Indeed, α-diversity measured with the Shannon diversity index increased significantly (p < 0.05) from 0.32 [<0.01–0.95] to 1.10 [<0.01–1.75] from NEFT-1 to NEFT-3 and from 0.64 [<0.01–1.28] to 1.39 [<0.01–1.89] from Me/F1 to F3, respectively.
The diversity and number of isolates obtained from those media specifically designed for the isolation of antibiotic-resistant bacteria are shown in Fig. 6. The number of isolates able to grow in the presence of methicillin (188 isolates) was six times higher than that isolated in the presence of vancomycin and carbapenems, whose numbers were similar (30 and 27 isolates, respectively). S. epidermidis (88 isolates) was the dominant species among methicillin-resistant isolates. Most of the vancomycin-resistant isolates belonged to the species Enterococcus faecalis and Enterobacter hormaechei (33% and 26%, respectively). Carbapenem-resistant isolates were detected in the samples collected on day 17 and some species of Enterobacter were more abundant in the NEFT samples than in the fecal ones. E. hormaechei (33%) was also the most abundant species among this group.
Distribution over time of the bacterial species isolated from NEFTs and fecal samples that grew on a MRSA Brilliance: MRSA 2 Agar for the isolation of methicillin-resistant bacteria, b VRE Brilliance: VRE Agar for the isolation of vancomycin-resistant bacteria , and c CRE Brilliance: CRE Agar for isolation of carbapenem-resistant bacteria.
Effect of type of feeding, antibiotherapy, and probiotic treatment in NEFTs and feces colonization
Potential associations between type of feeding, antibiotherapy, and probiotic treatment and the median (IQR) counts on the different culture media in NEFT-3 and F3 samples are included in Table 2. A similar analysis was performed for samples taken at times 1 and 2 and no statistically significant differences were obtained (Supplementary Table S7). Globally, the feeding type was not a significant driving factor neither in the bacterial profile of NEFT samples nor in the fecal ones, except for the Enterobacteriaceae counts in the case of the NEFT samples (p = 0.018). Antibiotic treatments had no impact on the median values of the bacterial counts, except for Pseudomonas spp. counts in NEFT samples (p = 0.01), where antibiotherapy was linked to an increase (>2 log10 CFU/cm2) in the median values. As for the probiotic treatment, 94% of the L. reuteri isolates (in total, 34) were recovered from those children whose feeding was supplemented with L. reuteri DSM 17938 (BioGaia ProTectis drops). Genotyping of these NEFTs and fecal isolates by random amplification polymorphism DNA showed that 53% (18) of the strains had the same profile as the commercial one (data not shown). When comparing the samples from infants with and without probiotic intervention, a concomitant reduction was observed in the median values of fecal MRSA and MRScys counts in the first group (p = 0.011 and 0.042, respectively).
Discussion
Microbial colonization of preterm infants is highly conditioned by prematurity itself but, also, by the circumstances that surround them during their NICU stay. In fact, the length of hospitalization is considered a factor that has a profound impact on the microbial colonization of the preterm infant.37 Since a high proportion of these infants are fed through NEFTs, these devices could serve as connectors between the environment and the intestinal microbiota. Therefore, the objective of this work was to study the potential parallelisms between NEFTs and fecal bacterial populations during the first 17 days from birth.
NEFT-1 showed a colonization profile different from the rest of the samples (and dominated by staphylococcal isolates) suggesting the first colonizers may arise from the feeding (OMM-fed neonates), the skin microbiota of the neonates, parents and/or hospital workers, and/or from the hospital environment. A relevant proportion (23%) of the NEFT-1 cultivable bacteria were isolated in the presence of methicillin, indicating that the impact of the hospital environment on preterm colonization starts immediately after birth as described in previous studies.37 On the contrary, other authors did not find antibiotic-resistant isolates in NEFTs samples during the first days of life despite almost half of them received antibiotherapy.10,38
The culture-based analysis of NEFT samples revealed a significant increase of the detection frequency and the bacterial load with time, except for vancomycin- and carbapenem-resistant microorganisms. In our study, NEFT-2 and NEFT-3 samples were mostly colonized by species belonging to the genera Staphylococcus and Enterococcus, and to the family Enterobacteriaceae. These results agree with the findings of previous studies in which these bacterial groups were dominant inside NEFTs.10,16,19,25,28 Their ability to form dense biofilms rapidly may provide them with a competitive advantage to colonize the inner surfaces of these devices.39 In addition, the lower gastric acidity that characterizes preterm neonates may also favor their growth in this environment.27
The role of NEFTs as reservoirs of potentially pathogenic microorganisms has been described in previous works.19,40 The inner surface of these devices is rather prone to be colonized by biofilm-forming microorganisms, which find optimal conditions for adhesion and proliferation, including a hydrophobic surface (silicone, PVC), constant supply of nutrients, and optimal growth temperature.41 Thick biofilm structures have already been observed within NEFTs, even 24 h after their insertion in the neonates.16 Probably, a relevant part of the bacterial load detected inside the feeding tubes in our study were biofilm-associated bacteria since the NEFTs remained 48 h inside the patients. Residues of the feedings provided through the NEFTs can positively influence the adhesion capacity of some microorganisms.16,29 In addition, the initial establishment of certain microorganisms, including Staphylococcus spp., on a surface may facilitate the adhesion of others, which will contact later with the same surface.42,43,44 All these features suggest that NEFTs can function as bidirectional routes for microbial exchange between the hospital environment and the infant gut.15
The fecal microbial pattern of NICU patients seems to be very similar among those neonates that share the same unit. In fact, the existence of a specific NICU-related microbiota has already been proposed14,15 and, overall, our data reinforce such a proposal and agree with previous works on the composition of the intestinal microbiota of preterm infants.16,45,46,47 In our study, staphylococci were dominant in the first samples but decreased over time, coinciding with the increase in the populations of enterococci and Enterobacteriaceae, which compete for nutrients and space to colonize the same biological niche. Similar shifts in the fecal cultivable bacteria after the first weeks of life have already been described for both preterm and term infants.30,48,49 Moreover, the probability of cross-transmission between patients hospitalized in the same NICU increases over time,50,51 which may partly explain why the bacterial profiles of NEFT-3 and F3 samples were more homogeneous than those observed in NEFT-1, Me, and F1, in which a higher degree of individual variability was observed.
The impact of feeding on the development of the intestinal microbiota of the newborn has been demonstrated in several works. In general, infants fed with OMM have a higher relative abundance in Lactobacillales and Clostridiales and more diversity than those who receive other types of feeding (formula or pasteurized DHM).52,53,54 In agreement with previous works,30,37 we observed that Enterobacteriaceae counts were higher among patients from the DHM group (who mostly received pasteurized donor milk) than in those from the OMM or MF groups. This suggests that some of the components of unpasteurized human milk (soluble immune factors, immune cells, bacteria or human milk oligosaccharides, among others) may participate in the shaping of the preterm gut microbiota.54,55,56 Currently, the actual impact of the feeding type in the colonization of preterm infants is relatively unknown since, in practice, it is very difficult to form groups of infants exclusively fed with OMM, exclusively fed with DHM, and exclusively fed with formula. Interestingly, feeding supplementation with the probiotic L. reuteri DSM 17938, a strain originally isolated from human milk, led to reduced values of methicillin-resistant counts, reinforcing the fact that some probiotic strains can stimulate the growth of lactobacilli and successfully compete with staphylococci.10,57,58 A concomitant reduction in Gram-positive counts (MRScys) was registered, probably due to the inhibition of the staphylococcal population that grows in this medium (approximately 10% of the staphylococci in this study were isolated from MRScys).
The administration of antibiotics to the preterm significantly increased the population of Pseudomonas spp. in the NEFTs, suggesting that antibiotherapy confers certain advantages to some specific Gram-negative bacteria that are intrinsically resistant to several antibiotics as previously reported in the case of Pseudomonas aeruginosa.59 Other studies have shown that antibiotics can greatly disrupt the diversity of the intestinal microbiota of preterm infants.38,60,61,62 In the short timespan covered by our study we did not observe such changes (at least in the microbial groups here analyzed) although 67% of the preterm infants in this study took antibiotics during their hospital stay. On the other hand, this approach only covers microorganisms that are cultivable under these conditions. Thus, we cannot discard other species that could be affected by these variables.
Interestingly, NEFTs and fecal samples of some of the infants harbored E. hormaechei isolates that exhibited resistance to methicillin in a sampling time, and resistance to vancomycin and carbapenems in the following sampling time. Further studies are required to elucidate if such isolates belong to different clones or if they belong to the same clone or strain that, as a consequence of the selective pressure, has acquired successive resistances through chromosomal mutations or through the acquisition of transmissible antibiotic resistance genes. Living within a biofilm, such as those formed within NEFTs, facilitates the exchange of genetic material through horizontal gene transference mechanisms,63 and, therefore, bacterial communities inside NEFTs could act as reservoirs of antibiotic resistance genes in the hospital setting.64,65
To summarize, this study demonstrates that the strong influence exerted by the hospital environment on the colonization of preterm infants starts immediately after birth. Our results show a parallel colonization of feces and NEFTs during the first 2 weeks of life, supporting the idea that there is an ongoing bidirectional transfer of microorganisms from the neonatal gut to the NEFT and vice versa. As NEFT populations seem to reflect the bacteria that are colonizing the preterm infants in a precise moment, their knowledge could be extremely useful to prevent the dissemination of antibiotic-resistant bacterial clones in the NICU and may guide future strategies to modulate the composition of the biofilm through the artificial creation of “healthier” NEFT bacterial populations before their use in neonates.
Limitations
The samples collected for this work were frozen at the hospital upon receipt and stored at −80 °C until processing. Under these conditions, those microorganisms especially sensitive to freeze–thaw cycles could have been not fully recovered. On the other hand, freezing is the most widely used method for the preservation of biological samples. Although we are conscious that this is a limitation of the study, the results here obtained represent a valuable contribution to the temporal evolution of the living bacterial communities that are colonizing the preterm complementing metataxonomic approaches in which the physiological state of the microorganisms is disregarded.
References
WHO. Preterm birth. https://www.who.int/en/news-room/fact-sheets/detail/preterm-birth (2018).
Howson, C. P., Kinney, M. V., McDougall, L. & Lawn, J. E. Born too soon: preterm birth matters. Reprod. Health 10, 1–9 (2013).
Moles, L. et al. Serratia marcescens colonization in preterm neonates during their neonatal intensive care unit stay. Antimicrob. Resist. Infect. Control 8, 1–8 (2019).
Arboleya, S. et al. Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol. Ecol. 79, 763–772 (2012).
Vinall, J. & Grunau, R. E. Impact of repeated procedural pain-related stress in infants born very preterm. Pediatr. Res. 75, 584–587 (2014).
Patel, K., Konduru, K., Patra, A. K., Chandel, D. S. & Panigrahi, P. Trends and determinants of gastric bacterial colonization of preterm neonates in a NICU setting. PLoS ONE 10, e0114664 (2015).
Henderickx, J. et al. The preterm gut microbiota: an inconspicuous challenge in nutritional neonatal care. Front. Cell Infect. Microbiol. 9, 85 (2019).
Jia, J. et al. Impact of postnatal antibiotics and parenteral nutrition on the gut microbiota in preterm infants during early life. J. Parent. Enter. Nutr. 44, 639–654 (2020).
Bu’Lock, F., Woolridge, M. W. & Baum, J. D. Development of co-ordination of sucking, swallowing and breathing: ultrasound study of term and preterm infants. Dev. Med. Child Neurol. 32, 669–678 (1990).
Petersen, S. M., Greisen, G. & Krogfelt, K. A. Nasogastric feeding tubes from a neonatal department yield high concentrations of potentially pathogenic bacteria- even 1 d after insertion. Pediatr. Res. 80, 395–400 (2016).
Berthelot, P. et al. Nosocomial colonization of premature babies with Klebsiella oxytoca: probable role of enteral feeding procedure in transmission and control of the outbreak with the use of gloves. Infect. Control Hosp. Epidemiol. 22, 148–151 (2001).
Martineau, C. et al. Serratia marcescens outbreak in a neonatal intensive care unit: new insights from next-generation sequencing applications. J. Clin. Microbiol. 56, e00235–18 (2018).
Young, B. E. et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA 323, 1488–1494 (2020).
Schwiertz, A. et al. Development of the intestinal bacterial composition in hospitalized preterm infants in comparison with breast-fed, full-term infants. Pediatr. Res. 54, 393–399 (2003).
Brooks, B. et al. Microbes in the neonatal intensive care unit resemble those found in the gut of premature infants. Microbiome 2, 1 (2014).
Gómez, M. et al. Early gut colonization of preterm infants: effect of enteral feeding tubes. J. Pediatr. Gastroenterol. Nutr. 62, 893–900 (2016).
Ogrodzki, P. et al. Rapid in situ imaging and whole genome sequencing of biofilm in neonatal feeding tubes: a clinical proof of concept. Sci. Rep. 7, 15948 (2017).
Donlan, R. M. Biofilms and device-associated infections. Emerg. Infect. Dis. 7, 277 (2001).
Mehall, J. R., Kite, C. A., Gilliam, C. H., Jackson, R. J. & Smith, S. D. Enteral feeding tubes are a reservoir for nosocomial antibiotic-resistant pathogens. J. Pediatr. Surg. 37, 1011–1012 (2002).
ECDC (European Centre for Disease Prevention and Control). Healthcare-associated infections acquired in intensive care units. ECDC. Annual epidemiological report for 2016. Stockholm: ECDC; 2018.
Hagbø, M. et al. Experimental support for multidrug resistance transfer potential in the preterm infant gut microbiota. Pediatr. Res. 88, 57–65 (2020).
Baldassarre, M. et al. Dysbiosis and prematurity: Is there a role for probiotics? Nutrients 11, 1273 (2019).
Høiby, N. et al. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin. Microbiol. Infect. 21, S1–S25 (2015).
Zingg, W. et al. Health-care-associated infections in neonates, children, and adolescents: an analysis of paediatric data from the European Centre for Disease Prevention and Control point-prevalence survey. Lancet Infect. Dis. 17, 381–389 (2017).
Hurrell, E. et al. Neonatal enteral feeding tubes as loci for colonisation by members of the Enterobacteriaceae. BMC Infect. Dis. 9, 146 (2009).
Poets, C. F. Gastroesophageal reflux: a critical review of its role in preterm infants. Pediatrics 113, e128–e132 (2004).
Segal, R., Pogoreliuk, I., Dan, M., Baumoehl, Y. & Leibovitz, A. Gastric microbiota in elderly patients fed via nasogastric tubes for prolonged periods. J. Hosp. Infect. 63, 79–83 (2006).
Mehall, J. R. et al. Prospective study of the incidence and complications of bacterial contamination of enteral feeding in neonates. J. Pediatr. Surg. 37, 1177–1182 (2002). (b).
Kim, H., Ryu, J. H. & Beuchat, L. R. Attachment of and biofilm formation by Enterobacter sakazakii on stainless steel and enteral feeding tubes. Appl. Environ. Microbiol. 72, 5846–5856 (2006).
Cong, X. et al. Gut microbiome developmental patterns in early life of preterm infants: impacts of feeding and gender. PLoS ONE 11, e0152751 (2016).
Mediano, P. et al. Microbial diversity in milk of women with mastitis: potential role of coagulase-negative Staphylococci, Viridans group Streptococci, and Corynebacteria. J. Hum. Lact. 33, 309–318 (2017).
Pohlert, T. PMCMRplus: calculate pairwise multiple comparisons of mean rank sums extended. R package version 1.9.0. https://CRAN.R-project.org/package=PMCMRplus (2021).
Hervé, M. RVAideMemoire: testing and plotting procedures for biostatistics. R package version 0.9-80. https://CRAN.R-project.org/package=RVAideMemoire (2021).
Mangiafico, S. rcompanion: functions to support extension education program evaluation. R package version 2.4.1. https://CRAN.R-project.org/package=rcompanion (2021).
Warnes, G. R. et al. gplots: various R programming tools for plotting data. R package version 3.1.1. https://CRAN.R-project.org/package=gplots (2020).
Oksanen, J. et al. Vegan: community ecology package. http://CRAN.R-project.org/package=vegan (2012).
Dahl, C. et al. Preterm infants have distinct microbiomes not explained by mode of delivery, breastfeeding duration or antibiotic exposure. Int. J. Epidemiol. 47, 1658–1669 (2018).
Gasparrini, A. J. et al. Antibiotic perturbation of the preterm infant gut microbiome and resistome. Gut Microbes 7, 443–449 (2016).
Liu, W. Y., Wong, C. F., Chung, K. M., Jiang, J. W. & Leung, F. C. Comparative genome analysis of Enterobacter cloacae. PLoS ONE 8, e74487 (2013).
Bussy, V., Marechal, F. & Nasca, S. Microbial contamination of enteral feeding tubes occurring during nutritional treatment. J. Parenter. Enter. Nutr. 16, 552–557 (1992).
Liu, R. et al. Molecular analysis of long-term biofilm formation on PVC and cast iron surfaces in drinking water distribution system. J. Environ. Sci. 26, 865–874 (2014).
Bur, S., Preissner, K. T., Herrmann, M. & Bischoff, M. The Staphylococcus aureus extracellular adherence protein promotes bacterial internalization by keratinocytes independent of fibronectin-binding proteins. J. Invest. Dermatol. 133, 2004–2012 (2013).
Puga, C. H., Dahdouh, E., SanJose, C. & Orgaz, B. Listeria monocytogenes colonizes Pseudomonas fluorescens biofilms and induces matrix over-production. Front. Microbiol. 9, 1706 (2018).
Jara, J. et al. Role of Lactobacillus biofilms in Listeria monocytogenes adhesion to glass surfaces. Int. J. Food Microbiol. 334, 108804 (2020).
Donnet-Hughes, A. et al. Potential role of the intestinal microbiota of the mother in neonatal immune education. Proc. Nutr. Soc. 69, 407–415 (2010).
Groer, M. W. et al. Development of the preterm infant gut microbiome: a research priority. Microbiome 2, 38 (2014).
Wang, Z. et al. Comparing gut microbiome in mothers’ own breast milk- and formula-fed moderate-late preterm infants. Front. Microbiol. 11, 891 (2020).
Jiménez, E. et al. Staphylococcus epidermidis: a differential trait of the fecal microbiota of breast-fed infants. BMC Microbiol. 8, 143 (2008).
Moles, L. et al. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS ONE 8, e66986 (2013).
Swartz, M. N. Hospital-acquired infections: diseases with increasingly limited therapies. Proc. Natl Acad. Sci. USA 91, 2420–2427 (1994).
de Man, P. et al. Enterobacter species in a pediatric hospital: horizontal transfer or selection in individual patients? J. Infect. Dis. 184, 211–214 (2001).
Gregory, K. E. et al. Influence of maternal breast milk ingestion on acquisition of the intestinal microbiome in preterm infants. Microbiome 4, 68 (2016).
Macpherson, A. J., de Agüero, M. G. & Ganal-Vonarburg, S. C. How nutrition and the maternal microbiota shape the neonatal immune system. Nat. Rev. Immunol. 17, 508–517 (2017).
Cong, X. et al. Influence of feeding type on gut microbiome development in hospitalized preterm infants. Nurs. Res. 66, 123–133 (2017).
Fernández, L., Ruiz, L., Jara, J., Orgaz, B. & Rodríguez, J. M. Strategies for the preservation, restoration and modulation of the human milk microbiota. implications for human milk banks and neonatal intensive care units. Front. Microbiol. 9, 2676 (2018).
Beghetti, I. et al. Human milk’s hidden gift: implications of the milk microbiome for preterm infants’ health. Nutrients 11, 2944 (2019).
Underwood, M. A. et al. A randomized placebo-controlled comparison of 2 prebiotic/probiotic combinations in preterm infants: impact on weight gain, intestinal microbiota, and fecal short-chain fatty acids. J. Pediatr. Gastroenterol. Nutr. 48, 216–225 (2009).
Esaiassen, E. et al. Effects of probiotic supplementation on the gut microbiota and antibiotic resistome development in preterm infants. Front. Pediatr. 6, 347 (2018).
Masuda, N. et al. Contribution of the MexX-MexY-OprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44, 2242–2246 (2000).
Jernberg, C., Löfmark, S., Edlund, C. & Jansson, J. K. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 1, 56–66 (2007).
Cox, L. M. et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158, 705–721 (2014).
Nobel, Y. R. et al. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat. Commun. 6, 7486 (2015).
Flemming, H. C. & Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633 (2010).
Fux, C. A., Costerton, J. W., Stewart, P. S. & Stoodley, P. Survival strategies of infectious biofilms. Trends Microbiol. 13, 34–40 (2005).
Sabater, S. et al. Monitoring the effect of chemicals on biological communities. The biofilm as an interface. Anal. Bioanal. Chem. 387, 1425–1434 (2007).
Acknowledgements
We thank the nursing team of the Neonatology Unit of La Paz University Hospital in Madrid (Spain) for their assistance and the Spanish Ministry of Economy and Competitiveness and the Spanish Pediatric Association Grant (2015) for their funding.
Funding information
This work has been funded by the Spanish Ministry of Economy and Competitiveness (project ref. PID2019-105606RB-I00) and by the Spanish Pediatric Association Grant (AEP, 2015).
Author information
Authors and Affiliations
Contributions
J.J.P. and I.C.N carried out the microbiological analysis of the samples and wrote the first draft of the manuscript. C.A.R. and L.F.A carried out the statistical analysis. B.M.- S. coordinated the clinical part of the study. B.M.-S, B.C.J., E.E.P., and M.S.P. enrolled infants and collected the clinical data. M.S.P., B.O.M., and J.M.R. conceived and designed the study. L.F.A. contributed to the study design. All authors contributed to the analysis, and/or interpretation of data. All authors revised the article critically and approved the final version to be published.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of La Paz University Hospital of Madrid, Spain (protocol code PI-3199 and date of approval, 27 June 2018). Parents or guardians provided written informed consent prior to enrollment.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jara Pérez, J., Moreno-Sanz, B., Castro Navarro, I. et al. Nasogastric enteral feeding tubes modulate preterm colonization in early life. Pediatr Res 92, 838–847 (2022). https://doi.org/10.1038/s41390-021-01852-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-021-01852-5