Abstract
Background
Fetal responses to adverse pregnancy environments are sex-specific. In fetal guinea pigs (GPs), we assessed morphology and messenger RNA (mRNA) expression in fetal growth-restricted (FGR) tissues at midpregnancy.
Methods
Female GPs were assigned either an ad libitum diet (C) or 30% restricted diet (R) prior to pregnancy to midpregnancy. At midpregnancy, a subset of R females underwent ultrasound-guided nanoparticle (NP) injection to enhance placental function. Five days later, fetuses were sampled. Fetal brain, heart, and liver were assessed for morphology (hematoxylin and eosin), proliferation (Ki67), and vascularization (CD31), as well as expression of inflammatory markers.
Results
R fetuses were 19% lighter with reduced organ weights and evidence of brain sparing compared to controls. No increased necrosis, proliferation, or vascularization was found between C and R nor male or female fetal organs. Sexual dimorphism in mRNA expression of Tgfβ and Ctgf was observed in R but not C fetal brains: increased expression in females. NP treatment increased fetal brain mRNA expression of Tgfβ and Ctgf in R males, abolishing the significant difference observed in untreated R fetuses.
Conclusions
Sex-specific differences in mRNA expression in the fetal brain with FGR could impart a potential survival bias and may be useful for the development of treatments for obstetric diseases.
Impact
-
Male and female fetuses respond differently to adverse pregnancy environments.
-
Under fetal growth restriction conditions, inflammatory marker mRNA expression in the fetal brain was higher in females compared to males.
-
Differences in gene expression between males and females may confer a selective advantage/disadvantage under adverse conditions.
-
Better characterization of sexual dimorphism in fetal development will aid better development of treatments for obstetric diseases.
Similar content being viewed by others
Introduction
Fetal growth restriction (FGR) is defined by the American College of Obstetricians and Gynecologists as estimated fetal growth for gestational age ≤10th percentile with a live birth rate incidence of 4–8% in developed countries and 6–30% in developing countries.1 FGR is a common cause of medically indicated iatrogenic preterm birth, and the second most common cause of neonatal morbidity and mortality behind spontaneous preterm birth, placing a significant burden on health-care systems and social costs related to long-term disease and disability.2 The development of FGR is still largely unknown; however, maternal, fetal, and placental characteristics have all been implicated as causative factors. Most abundantly studied is uteroplacental insufficiency, where the impaired exchange of nutrients and oxygen at the placental interface does not allow the fetus to reach its full growth potential.3 However, other environmental factors such as a maternal diet, pollution, substance abuse, and chronic stress are all known to be associated with increasing the risk of FGR.4
Another factor affecting fetal growth in utero is fetal sex. It is well established that fetal sex influences fetal and neonatal morbidity and mortality in humans,5 as well as animal models.6 In general, male fetuses experience poorer pregnancy outcomes including increased incidences of preterm delivery and stillbirth.5 This is thought to have evolutionary origins where males are traditionally viewed as the more “costly” sex and the maternal system is less likely to invest long term in the development of a male under adverse maternal conditions.7,8 Furthermore, female fetuses appear to have more placental adaptations to adverse pregnancy outcomes suggesting an innate ability to adapt to stressors and thus more likely to result in a favorable pregnancy outcome.9,10 Sex-specific differences in placental development and function have been studied;11 however, the mechanistic link between adverse maternal conditions and the placental/fetal response is poorly understood.
Guinea pigs offer a unique advantage in the study of adverse pregnancy outcomes and particular FGR via noninvasive maternal nutrient restriction (MNR). In humans, epidemiologic study of the Dutch famine during World War II showed that a major reduction in caloric uptake equivalent to 75% food restriction during pregnancy was needed to produce significant fetal weight reduction by >300 g in human infants.12 In guinea pigs, MNR of 30%, 4 weeks prior to conception and until midpregnancy, and then 10% thereafter, significantly reduces fetal body weight.13 Fetal body composition is also altered in FGR offspring with skeletal muscle, retroperitoneal adiposity, thymus, and spleen decreased; heart, kidneys, and adrenals remained the same; while brain and lungs increased relative to fetal body weight indicating a pattern of blood redistribution for fetal brain sparing.14 Furthermore, this model does not result in extreme fetal loss and only modest increased preterm delivery rate,15 overall validating this model for translational study. The aim of the current study was to understand the impact of MNR on the developing fetal organs in midpregnancy and assess potential sexual dimorphisms that could aid future studies developing therapeutics for placental dysfunction and FGR.
Methods
Animal care and usage
Animal care, mating, and surgical procedures were approved by the Institutional Animal Care and Use Committee at Cincinnati Children’s Hospital Medical Center and Research Center (Protocol number 2017-0065). Female Dunkin–Hartley guinea pigs were purchased (Charles River Laboratories, Wilmington, MA) at ~6–8 weeks of life and housed in a temperature-controlled environment with a 12 h light–dark cycle and placed on ad libitum chow (LabDiet diet 5025: 27% protein, 135 fat, and 60% carbohydrate as % of energy) with tap water ad libitum during their acclamation.
Following a 2-week acclimation, females were weighed and assigned into two groups: control (C), n = 7 or MNR/R, n = 5. In the R group, food intake was restricted to 70% of the C group for 4 weeks preconception through midpregnancy (gestational day (GD) 30). To prevent pregnancy wastage, intake was increased to 90% of C at GD30. Throughout the 4-week dietary acclimation period, the estrus cycle was monitored daily by observing the perforation of the vaginal membrane.16 Females were placed with males overnight when the vaginal membrane was observed as beginning to perforate, and remained with the males during the nighttime period only until closure of the vaginal membrane. Ovulation was assumed to occur when the vaginal membrane was fully perforated and designated GD1. These animals were used as part of a larger study to determine the effects of nanoparticle (NP) treatment on placental function and therefore underwent an ultrasound-guided, trans-uterine, intra-placental injection of 200 µL phosphate-buffered saline at GD30–33. A second cohort of R females (R-NP): n = 7, received an ultrasound-guided trans-uterine intra-placental injection of NP (240 µg plasmid in 200 µL injection). This NP was composed of a plasmid containing the human insulin-like 1 growth factor (hIGF1) gene under the control of a placenta-specific promoter (Cyp19a1) complexed with a nonviral, biodegradable HPMA-DMEAMA (N-(2-hydroxypropyl)methacrylamide-2-(dimethylamino)ethyl methacrylate) copolymer, which functions to protect the plasmid from degradation as well as to aide cellular uptake.17,18 All females were sacrificed 5 days after injection between GD35–38. At the time of sacrifice, dam was fasted for 4 h, weighed, and euthanized using carbon dioxide asphyxiation; the second method was maternal cardiac puncture with exsanguination. The pregnant uterine horns were dissected out and fetuses weighed. Fetal sex was determined based on examination of the fetal gonads and validated using polymerase chain reaction (PCR). Fetal organs including brain, heart, liver, and lungs were dissected and weighted. The dissected tissues were then divided in half and either placed in tissue cassettes and fixed in 4% w/v paraformaldehyde (PFA) for histology or flash frozen in liquid nitrogen and stored in −80 °C for later RNA extractions.
Fetal sex determination
Fetal sex determination at the time of necropsy was validated using PCR following the protocol outlined in ref. 19. Briefly, fetal ear was digested and genomic DNA (gDNA) extracted using the Qiagen gDNA Extraction Kit following standard protocols (Qiagen). PCR reactions were performed with primers that amplified guinea pig Dystrophin (Dys) and Sry with sequences as follows:19
Dys F: GTGTTAATGGTGACAGCATCAGC, R: TGCTGTTGGATCTGAAGTGGAGG;
Sry F: CCATGATTGCATTTATGGTGTGGTCCCG, R: GCCTTTTTTCGGCTTCTGTAAGCATTTTCCAC.
In 25 µL PCR reactions, 10 ng of fetal gDNA was mixed with 12.5 µL of EconoTaq DNA polymerase (Lucigen), 0.4 µM of each Dys primer and 0.8 µM of each Sry primer. PCR cycling then proceeded as follows: 95 °C for 3 min, 36 cycles at 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s, followed by a final cycle at 72 °C for 5 min. PCR products were then run on a 10% TBE (Tris/Borate/EDTA) gel (Invitrogen) and stained with GelRed nucleic acid gel stain (Biotium) to visualize bands. gDNA from known female and male origins were also included as reference samples.
Morphological analysis and immunohistochemistry
For morphological analysis of fetal heart, lung, and brain, 5-µm-thick fixed tissue sections were cut using a microtome. Hematoxylin and eosin staining was used following standard laboratory protocol to assess gross tissue morphology. Immunohistochemistry (IHC) was undertaken to show CD31, an endothelial cytoplasm marker, and Ki67, a nuclear proliferation marker expression. The 5 µm sections were deparaffinised and rehydrated following standard protocol. Antigen retrieval was performed by incubating slides in 10× Target Retrieval Solution (Dako) for 15–30 min in a bead bath at 95 °C and then cooled at room temperature. Endogenous peroxidase activity was suppressed by incubating the slides in 3% hydrogen peroxide and then a protein block of fish skin gelatin, 5% goat serum, and 1% bovine serum albumin was applied to the sections for 1 h at room temperature. Primary antibodies for CD31 (Abcam ab28364; diluted 1:50) and Ki67 (Invitrogen MA5-14520; diluted 1:200) were diluted in the protein block and applied overnight at 4 °C under humidified conditions. Negative controls were included by omitting the primary antibody from the diluent. CD31 and Ki67 antibody binding was amplified with goat anti-rabbit secondary antibody (Invitrogen; diluted 1:200), followed by incubation with ABC reagents (Vector) and visualized using 3,3′-diaminobenzidine (DAB) (Vector). Nuclei were counterstained with hematoxylin and sections set as previously described. Final images of the staining were captured using the Nikon Eclipse 80i light microscope. For analysis of Ki67, ten images per section at ×40 magnification were captured and cell counts were performed using the automated cell counting function in the Nis Elements Basic Research software (Nikon). Both positive DAB-stained nuclei and negative hematoxylin stained nuclei were counted. For analysis of CD31, a similar approach was used except the number of blood vessels per field counted manually.
Fetal brain in situ hybridization
To confirm the inability of the NP to cross into fetal circulation, in situ hybridization was used to determine plasmid-specific messenger RNA (mRNA) expression in representative fetal brain sections. Using a custom-designed BaseScopeTM probe (Advanced Cell Diagnostics) specific to the sequence between the stop codon and polyA signaling of the plasmid, 5-µm-thick PFA-fixed, paraffin-embedded tissue sections were deparaffinized and rehydrated following protocols outlined in the standard BaseScopeTM protocol. Extended antigen retrieval was then performed by boiling for 30 min in RNAscopeTM Targeted Retrieval Reagent (Advanced Cell Diagnostics), followed by incubation with RNAscopeTM Protease IV (Advanced Cell Diagnostics) for 30 min at 40 °C. In situ hybridization with the BaseScopeTM assay to visualize plasmid-specific mRNA occurred following the manufacturer’s protocol and sections were counterstained with hematoxylin. Imaging occurred using the Nikon Eclipse 80i microscope and a section of NP-injected guinea pig placenta was used as a positive control.
RNA isolations, reverse transcription, and quantitative PCR (qPCR)
For RNA extractions, flash-frozen fetal heart, liver, and brain samples were homogenized in RLT lysis buffer (Qiagen) with 1% β-mercaptoethanol and agitation was aided by a tissue homogenizer. Total cellular RNA was isolated using the RNeasy Mini RNA Isolation Kit (Qiagen) as per the manufacturer’s protocol. RNA quantification was assessed using Nanodrop® Spectrophotometer (Thermo Fisher). 1 µg of total RNA was used to generate complementary DNA (cDNA) using the High Capacity cDNA Reverse Transcription Kit following the manufacturer’s protocol (Applied Biosystems). Inflammatory response markers were quantified qPCR with primers against transforming growth factor β (Tgfβ), connective tissue growth factor (Ctgf), tissue necrosis factor-α (Tnfα), matrix metalloproteinase 2 (Mmp2), and normalized to the housekeeping gene β-actin (β-Actin) as follows:
Tgfβ F: CAATTCCTGGCGCTACCTCA, R: ACCGATCCGTTGATTTCC;
Ctgf F: CACCCGGGTTACCAATGACA, R: CCGGTAGGTCTTCATGCTGG;
Tnfα F: GCCGTCTCCTACCCGGAAAA, R: TAGATCTGCCCGGAATCGGC;
Mmp2 F: CAGGGCACCTCCTACAACAG, R: CCTCTGAGTCCCCACCGAC;
β-Actin F: CGCGAGAAGATGACCCAG, R: TAGCACAGCCTGGATAGCAA.
qPCR analysis was performed in 20 µL triplicate reactions containing Power SYBR Green Master Mix (Invitrogen), 600 nM of primers, and 2.5 µL cDNA by the Applied Biosystems StepOne-Plus Real-Time PCR System. Relative mRNA expression was calculated using the StepOne Software v2.3 (Applied Biosystems) by the comparative CT method.
Statistical analysis
All data were analyzed using generalized linear modeling in SPSS Statistics Software (v.26). Each variable was adjusted for gestational age; fetal sex and maternal diet were considered main effects, while the random effect of maternal in utero environment was also included. Post hoc analyses were performed using Bonferroni adjustment. For additional analyses including the R-NP group, NP treatment was also included within the model as the main effect. Data are reported as the estimated marginal mean and standard error (SE), and statistical significance was deemed at P < 0.05.
Results
Maternal nutrient restriction induces FGR without pregnancy wastage
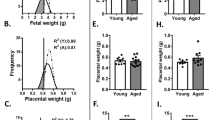
In the current study, MNR of female guinea pigs through to GD35–38 resulted in no fetal loss related to MNR and no significant difference in litter size between the C or R groups (C: 3.29 ± 0.52 vs. R: 2.80 ± 0.84; P > 0.05). There was also no difference in the proportions of female to male fetuses between the C and R diet groups (C female: 11 vs. C male: 10 vs. R female: 7 vs. R male: 7; Fisher’s test P = 0.721). While there was no difference in maternal carcass weight (maternal weight minus total fetal and placental weight) between C and R dams (Fig. 1a), fetal weight was decreased in the R group (Fig. 1b). There was no difference in placental weight (Fig. 1c) nor was there a difference between the fetal sexes for fetal and placental weight. Fetal heart, lung, and liver weight as a percentage of fetal body weight was lower in the R fetuses compared to C and was also not different between the sexes (Fig. 2a–c). Fetal brain weight as a percentage of body weight was, however, not different between the C and R fetuses (Fig. 3A), and the brain-to-liver weight ratio was increased in the R fetuses indicating brain sparing consistent with the physiologic blood redistribution typically seen in asymmetric FGR and not different between the sexes (Fig. 3b).
At midpregnancy, there was no difference in maternal weight (a) between control-fed and restricted-fed females. However, fetal weight (b) was reduced in the restricted-fed females, while placental weight remained similar between the diet groups (c). There was no difference between the fetal sexes for neither fetal nor placental weight. Data are estimated marginal mean ± SE, n = 7 control (11 female and 10 male fetuses) and n = 5 restricted (7 female and 7 male fetuses). Statistical significance was determined using generalized linear mixed modeling including diet and fetal sex as main effects, maternal in utero environment as a random effect, and gestational day as a covariate.
In fetuses from restricted-fed females, % of body weight of fetal heart (a), fetal lung (b), and fetal liver (c) was reduced compared to fetuses from control-fed females. For all organs, there was no difference between the fetal sexes. Data are estimated as marginal mean ± SE, n = 7 control (11 female and 10 male fetuses) and n = 5 restricted (7 female and 7 male fetuses). Statistical significance determined using generalized linear mixed modeling including diet and fetal sex as main effects, maternal in utero environment as a random effect, and gestational day as a covariate.
In the fetuses from restricted-fed females, brain weight as a % of body weight was similar to fetuses from control-fed females (a), while there was an increase in the brain-to-liver weight ratio (b) indicating brain sparring. There was no difference between fetal sexes. Data are estimated as marginal mean ± SE, n = 7 control (11 female and 10 male fetuses) and n = 5 restricted (7 female and 7 male fetuses). Statistical significance determined using generalized linear mixed modeling including diet and fetal sex as main effects, maternal in utero environment as a random effect, and gestational day as a covariate.
Histological analysis of tissue morphology and cell proliferation in fetal brain, heart, and liver
With smaller fetal weight and organ size, as well as brain sparing in the FGR fetuses, we sought to further investigate tissue morphology. Hematoxylin and eosin staining of the fetal brain, heart, and liver showed no evidence of increased tissue necrosis or inflammatory infiltration in the R tissue when compared to C fetuses (Fig. 4). IHC staining for Ki67 in the fetal brain revealed regions of high amounts of proliferation including the left ventricle and cerebellum (Fig. 5a, b). Interestingly, despite reduced organ weights, the percentage of cells positive for Ki67 in both fetal heart and liver was similar between C and R fetuses (Fig. 5c, d). There was also no difference in the percentage of cells positive for Ki67 in fetal heart or liver between males and females. IHC staining for CD31 to identify blood vessels in fetal brain and heart indicated no differences in blood vessel density with neither dietary intervention nor fetal sex (Supplementary Fig. 1).
Ki67 staining showed high levels of proliferation within the fetal brain, particularly in the left ventricle (closed arrow) and cerebellum (open arrow) regions of both control (a) and restricted (b) fetuses. The percentage of cells positive for Ki67 in the fetal heart was not different between control and restricted fetuses nor between male and female fetuses (c). Similarly, there was no difference in the percentage of cells positive for Ki67 in fetal liver between different diet groups or fetal sex (d). a and b are representative images, scale bar = 5 mm. Data in c and d are estimated marginal mean ± SE, n = 4 control (2 female and 4 male fetuses) and n = 3 restricted (3 female and 3 male fetuses).
Fetal brain mRNA expression of inflammatory markers Tgfβ and Ctgf differ with FGR and fetal sex
After recapitulating the FGR model, we then began to characterize some of the baseline characteristics of fetal programming in organs affected by blood redistribution in asymmetric FGR: fetal brain, heart, and liver. While no significant difference in mRNA expression of Tgfβ, Ctgf, Tnfα, or Mmp2 was found in fetal heart or liver (Supplementary Table 1), there was differences in fetal brain mRNA expression of Ctgf and Tgfβ. Ctgf and Tgfβ mRNA in the fetal brain was not different between female and male fetuses in the C dietary group; however, the expression was increased in female fetuses compared to male fetuses in the R dietary group (Fig. 6a, b). There was no difference in fetal brain expression of Tnfα nor Mmp2 between C and R fetuses nor between male and female fetuses (Fig. 6c, d).
There was no effect of fetal growth restriction due to maternal nutrient restriction on fetal brain mRNA expression of transforming growth factor β (Tgfβ, a), connective tissue growth factor (Ctgf, b), tissue necrosis factor-α (Tnfα, c), or matrix metalloproteinase 2 (Mmp2, d) when compared to control fetuses. However, fetal brain mRNA expression of Tgfβ was lower in the restricted male fetuses compared to restricted female fetuses (a). mRNA expression of Ctgf in the fetal brain was also lower in restricted males compared to restricted females (b). Data are estimated marginal mean ± SE, n = 6 control (11 female and 7 male fetuses) and n = 5 restricted (7 female and 7 male fetuses). Statistical significance was determined using generalized linear mixed modeling including diet and fetal sex as main effects, maternal in utero environment as a random effect, and gestational day as a covariate. Bonferroni post hoc analyses were also performed. *P < 0.05; ns not significant.
Placental treatment with NP modulates brain and fetal mRNA expression in a sex-specific manner
Placental treatment with NP has previously been shown to maintain fetal growth in a mouse model of surgically induced FGR.18 R-NP fetuses had reduced fetal weight compared to control and was similar to R fetal weight (Fig. 7a); placental weight was similar across all groups (Fig. 7b). No plasmid-specific mRNA was found in fetal brain tissue from R-NP treated fetuses confirming the inability of the NP to enter fetal circulation via the placenta (Fig. 7c). Of most interest, despite only directly affecting the placenta, NP treatment resulted in changes in fetal brain mRNA expression of both Tgfβ and Ctgf so that expression was similar between male and female fetuses in the R-NP group (Fig. 7d, e).
In growth-restricted fetuses, placental injection of nanoparticle (NP) to treat placental dysfunction did not change fetal weight when compared to untreated fetuses (a). Placental weight remained similar in nanoparticle-treated restricted females compared to untreated, restricted, and control fed (b). In situ hybridization of plasmid-specific mRNA confirmed the inability for the nanoparticle plasmid to enter fetal circulation as no plasmid-specific mRNA was found in the fetal brain of fetuses, which received nanoparticle treatment (c). However, nanoparticle treatment resulted in changes to fetal brain expression of transforming growth factor-β (Tgfβ, d) and connective tissue growth factor (Ctgf, e). Expression of both markers was lower in untreated restricted male fetuses compared to untreated restricted female fetuses. Data are estimated marginal mean ± SE, n = 7 control (11 female and 10 male fetuses) and n = 5 restricted (7 female and 7 male fetuses). Statistical significance was determined using generalized linear mixed-modeling including diet and fetal sex as main effects, maternal in utero environment as a random effect, and gestational day as a covariate. Bonferroni post hoc analyses were also performed. A and B indicate statistical significance of P < 0.05 between diet groups. *P < 0.05, **P < 0.01; ns not significant.
Discussion
There is continuing interest in identifying biomarkers for adverse pregnancy outcomes as this information can not only be used to better diagnose obstetric diseases but also as potential targets for therapeutics. Here, we were able to successfully induce FGR with asymmetrical fetal growth patterning resulting in brain sparing at midpregnancy in the guinea pig. Analysis of the key organs involved in blood redistribution: fetal heart, liver, and brain revealed for the first time sex-specific differences in brain expression of growth factors and inflammatory cytokines, which may confer a survival advantage or disadvantage as the pregnancy progresses. Furthermore, we have shown that treatment of the placenta with a NP, known to modulate fetal growth in other models of fetal growth restriction18,20 by increasing nutrient transporter expression and placental angiogenesis, can affect brain mRNA expression in sex-specific manners. Overall, such observations provide crucial understandings of fetal programming in FGR, which will inform future studies aimed at predicting and treating placental dysfunction in utero.
In the present study, we were able to successfully establish the guinea pig model of FGR through MNR as previously described.13,14,15 MNR resulted in reduced overall fetal weight at midpregnancy as well as reductions in key fetal organ weight, except for the fetal brain. Furthermore, our study successfully recapitulated the FGR model without impaired fertility, pregnancy wastage, similar fertility rates, and litter sizes, as well as no differences in the distribution of male and female pups. In humans, fetal growth occurs in three distinct phases during gestation with the second phase of growth in midpregnancy characterized by both rapid cellular proliferation (hyperplasia) and increased cellular size (hypertrophy).2 Perturbations to growth patterning during this time are predicted to result in asymmetric growth patterning with normal skeletal size but low fetal weight and is the hallmark of the 80% of FGR attributable to uteroplacental insufficiency. Current clinical practices, however, are more commonly incorporating the use of sonographic fetal biometry measures including umbilical artery Doppler to increase the predictive accuracy of diagnosing growth restriction in utero.21 Such techniques are only just being explored in the guinea pig MNR model with reduced biparietal diameter and head circumference observed as early as GD39.22 While these experiments are beyond the scope of the current study, they do present promising future directions that build up our current study, which highlights the usefulness of this model in continuing to elucidate the mechanistic causes and potential treatments of FGR.
Another key finding within the guinea pig MNR model was brain sparing. Organ sparing in FGR occurs due to blood redistribution from peripheral organs including the liver to essential organs such as the brain and heart based on organ size.23 Physiologically this is via the carotid chemoreflex and baroreceptors utilizing nitric oxide and adenosine to alter the vascular tone.24 However, histological analysis of fetal organs involved in blood redistribution in FGR: the brain, heart, and liver, showed no evidence of decreased nuclear hyperplasia or vascularization that would account for smaller fetal organ size or blood redistribution in midpregnancy. On the other hand, there was no evidence of increased cellular necrosis or increased inflammatory infiltrate in the organs of the restricted fetuses. While these preliminary investigations do not provide definitive conclusions on the biological mechanisms as to how fetal brain sparing is occurring in this model, such data does contribute necessary baseline measurements for further studies. These potentially include assessing glucose transporter 1, erythropoietin, and vascular endothelial growth factor to further evaluate fetal hypoxia–ischemia and neovascularization.25
The most compelling outcomes observed in this study were differences in mRNA expression of inflammatory markers between female and males fetuses. To our knowledge, we are the first to explore sexual dimorphism in the expression of Tgfβ, Ctgf, Tnfα, and Mmp2 in fetal liver, heart, and brain, comparing normal and growth-restricted fetuses. In male growth-restricted fetuses, brain mRNA expression of Ctgf and Tgfβ were significantly lower compared to female fetuses. It is well established that female and male fetuses respond differently to adverse obstetric environments.5 Furthermore, sexual dimorphism among FGR fetuses in the MNR guinea pig model have been previously shown near term.26 Increased hypoxyprobe-1 (a marker of chronic tissue hypoxia) in the fetal liver and kidneys was observed in all pups, but only female pups showed increased erythropoietin and vascular endothelial growth factor, which is hypothesized to impart a survival advantage during suboptimal uterine conditions.26 The impact of fetal sex on fetal outcome is also not restricted to models of maternal undernutrition. In studies of maternal obesity or high-fat diet animal models, there are known differences in placental function depending on fetal sex.27,28,29 Thus, these findings, that there are differences in expression of the inflammatory markers in males compared to females, which may provide a survival advantage/disadvantage, may also occur in other models of pregnancy perturbations and help reinforce the idea that sex-specific differences must be considered in all pregnancy studies.
Consideration of fetal sex in the development of pregnancy therapeutics is also important. When the placental function was modified through NP treatment, this resulted in increased mRNA expression in the fetal brain of the restricted males, abolishing the prior significant difference with restricted female fetuses. Further still, we showed the inability of our NP to cross the placental barrier and enter the fetus, and thus any differences in fetal mRNA expression were due to signaling events from the placenta. We did not observe any significant changes to fetal weight with NP treatment, although this was not unexpected as 5 days is a relatively short period of time when considering guinea pigs having a 65–70 day gestation. Nevertheless, the observed changes in gene expression in the present study may be important to the initial processes of restoring fetal growth. Without further studies, these mechanistic implications are unclear; however, these data have established a baseline understanding of these markers in fetal liver, heart, and brain, as well as shown the ability to modulate fetal gene expression, which will be necessary for future investigations.
In conclusion, we have shown for the first time sexual dimorphism in mRNA expression of a number of inflammatory markers in the fetal brain at midpregnancy in the guinea pig. Additionally, we have demonstrated that treatment of the placenta with a nanoparticle that enhances placental growth factor expression can differentially affect fetal organ mRNA expression depending on fetal sex. Such observations may impart a potential survival bias between females and males, particularly in adverse conditions. Overall, this study provides essential baseline data for further studies focused on not only better understanding the developmental origins of health and disease but also on the development of potential therapeutics to treat obstetric diseases.
References
Resnik, R. & Lockwood, C. Fetal growth restriction: Evaluation and management. UpToDate, Waltham, MA (Accessed Oct. 20, 2016). (2013).
Greene, M. F. et al. Creasy and Resnik’s Maternal-Fetal Medicine: Principles and Practice E-Book (Elsevier Health Sciences, 2008).
Baschat, A. A. & Hecher, K. Fetal growth restriction due to placental disease. Semin. Perinatol. 28, 67–80 (2004).
Roberts, C. T. IFPA Award in Placentology Lecture: complicated interactions between genes and the environment in placentation, pregnancy outcome and long term health. Placenta 31, S47–S53 (2010).
Vatten, L. J. & Skjaerven, R. Offspring sex and pregnancy outcome by length of gestation. Early Hum. Dev. 76, 47–54 (2004).
Aiken, C. E. & Ozanne, S. E. Sex differences in developmental programming models. Reproduction 145, R1–R13 (2013).
Rickard, I. J., Russell, A. F. & Lummaa, V. Producing sons reduces lifetime reproductive success of subsequent offspring in pre-industrial Finns. Proc. Biol. Sci. 274, 2981–2988 (2007).
Mathews, F., Johnson, P. J. & Neil, A. You are what your mother eats: evidence for maternal preconception diet influencing foetal sex in humans. Proc. Biol. Sci. 275, 1661–1668 (2008).
Vickers, M. H., Clayton, Z. E., Yap, C. & Sloboda, D. M. Maternal fructose intake during pregnancy and lactation alters placental growth and leads to sex-specific changes in fetal and neonatal endocrine function. Endocrinology 152, 1378–1387 (2011).
Wilcoxon, J. S., Schwartz, J., Aird, F. & Redei, E. E. Sexually dimorphic effects of maternal alcohol intake and adrenalectomy on left ventricular hypertrophy in rat offspring. Am. J. Physiol. Endocrinol. Metab. 285, E31–E39 (2003).
Clifton, V. L. Review: Sex and the human placenta: mediating differential strategies of fetal growth and survival. Placenta 31, S33–S39 (2010).
Lumey, L. H. & Stein, A. D. Offspring birth weights after maternal intrauterine undernutrition: a comparison within sibships. Am. J. Epidemiol. 146, 810–819 (1997).
Sohlstrom, A. et al. Food restriction alters pregnancy-associated changes in IGF and IGFBP in the guinea pig. Am. J. Physiol. 274, E410–E416 (1998).
Kind, K. L. et al. Chronic maternal feed restriction impairs growth but increases adiposity of the fetal guinea pig. Am. J. Physiol. Regul. Integr. Comp. Physiol. 288, R119–R126 (2005).
Elias, A. A., Ghaly, A., Matushewski, B., Regnault, T. R. & Richardson, B. S. Maternal nutrient restriction in guinea pigs as an animal model for inducing fetal growth restriction. Reprod. Sci. 23, 219–227 (2016).
Wilson, R., Lampe, K., Matushewski, B. J., Regnault, T. R. & Jones H. N. Time mating guinea pigs by monitoring changes to the vaginal membrane throughout the estrus cycle and with ultrasound confirmation. bioRxiv https://doi.org/10.1101/2020.08.21.256255 (2020).
Wilson, R. L. et al. Nanoparticle mediated increased insulin-like growth factor 1 expression enhances human placenta syncytium function. Placenta 93, 1–7 (2020).
Abd Ellah, N. et al. Development of non-viral, trophoblast-specific gene delivery for placental therapy. PLoS ONE 10, e0140879 (2015).
Depreux, F. F., Czech, L. & Whitlon, D. S. Sex genotyping of archival fixed and immunolabeled guinea pig cochleas. Sci. Rep. 8, 5156 (2018).
Wilson R. L. et al. Insulin-like growth factor 1 signaling in the placenta requires endothelial nitric oxide synthase to support trophoblast function and normal fetal growth. bioRxiv https://doi.org/10.1101/2020.08.21.261420 (2020).
Hiersch, L. & Melamed, N. Fetal growth velocity and body proportion in the assessment of growth. Am. J. Obstet. Gynecol. 218, S700–S711.e701 (2018).
Swanson, A. M. et al. The use of ultrasound to assess fetal growth in a guinea pig model of fetal growth restriction. Lab Anim. 51, 181–190 (2017).
Cahill, L. S., Zhou, Y. Q., Seed, M., Macgowan, C. K. & Sled, J. G. Brain sparing in fetal mice: BOLD MRI and Doppler ultrasound show blood redistribution during hypoxia. J. Cereb. Blood Flow Metab. 34, 1082–1088 (2014).
Morrison, S., Gardner, D. S., Fletcher, A. J., Bloomfield, M. R. & Giussani, D. A. Enhanced nitric oxide activity offsets peripheral vasoconstriction during acute hypoxaemia via chemoreflex and adrenomedullary actions in the sheep fetus. J. Physiol. 547, 283–291 (2003).
Li, Y., Gonzalez, P. & Zhang, L. Fetal stress and programming of hypoxic/ischemic-sensitive phenotype in the neonatal brain: mechanisms and possible interventions. Prog. Neurobiol. 98, 145–165 (2012).
Elias, A. A. et al. Maternal nutrient restriction in guinea pigs leads to fetal growth restriction with evidence for chronic hypoxia. Pediatr. Res. 82, 141–147 (2017).
Reynolds, C. M., Vickers, M. H., Harrison, C. J., Segovia, S. A. & Gray C. Maternal high fat and/or salt consumption induces sex-specific inflammatory and nutrient transport in the rat placenta. Physiol. Rep. 3, e12399 (2015).
Gabory, A. et al. Maternal diets trigger sex-specific divergent trajectories of gene expression and epigenetic systems in mouse placenta. PLoS ONE 7, e47986 (2012).
Evans, L. & Myatt, L. Sexual dimorphism in the effect of maternal obesity on antioxidant defense mechanisms in the human placenta. Placenta 51, 64–69 (2017).
Acknowledgements
We would like to thank Dr. Timothy Regnault at the University of Western Ontario for his assistance with designing the inflammatory cytokine oligonucleotide primer sequences as well as the staff at Vet Services, Cincinnati Children’s Hospital and Medical Center for their assistance with the animal procedures. This study was funded by Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) award R01HD090657 (H.N.J.).
Author information
Authors and Affiliations
Contributions
R.L.W. conceived the study, performed experiments, analyzed the data, and wrote the manuscript. K.K.S. conceived the study, performed experiments, analyzed the data, and wrote the manuscript. K.L. performed experiments and edited the manuscript. H.N.J. conceived the study, obtained the funding, and edited the manuscript. All authors approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Wilson, R.L., Stephens, K.K., Lampe, K. et al. Sexual dimorphisms in brain gene expression in the growth-restricted guinea pig can be modulated with intra-placental therapy. Pediatr Res 89, 1673–1680 (2021). https://doi.org/10.1038/s41390-021-01362-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-021-01362-4
This article is cited by
-
A Low Dose of Ouabain Alters the Metabolic Profile of Adult Rats Experiencing Intrauterine Growth Restriction in a Sex-Specific Manner
Reproductive Sciences (2023)
-
Working towards precision medicine in developmental programming
Pediatric Research (2021)