Abstract
Background
Bronchopulmonary dysplasia remains one of the most common complications of prematurity, despite significant improvements in perinatal care. Functional modeling of human lung development and disease, like BPD, is limited by our ability to access the lung and to maintain relevant progenitor cell populations in culture.
Methods
We supplemented Rho/SMAD signaling inhibition with mTOR inhibition to generate epithelial basal cell-like cell lines from tracheal aspirates of neonates.
Results
Single-cell RNA-sequencing confirmed the presence of epithelial cells in tracheal aspirates obtained from intubated neonates. Using Rho/SMAD/mTOR triple signaling inhibition, neonatal tracheal aspirate-derived (nTAD) basal cell-like cells can be expanded long term and retain the ability to differentiate into pseudostratified airway epithelium.
Conclusions
Our data demonstrate that neonatal tracheal aspirate-derived epithelial cells can provide a novel ex vivo human cellular model to study neonatal lung development and disease.
Impact
-
Airway epithelial basal cell-like cell lines were derived from human neonatal tracheal aspirates.
-
mTOR inhibition significantly extends in vitro proliferation of neonatal tracheal aspirate-derived basal cell-like cells (nTAD BCCs).
-
nTAD BCCs can be differentiated into functional airway epithelium.
-
nTAD BCCs provide a novel model to investigate perinatal lung development and diseases.
Similar content being viewed by others
Introduction
Bronchopulmonary dysplasia (BPD) remains the most frequent adverse outcome for infants born less than 30 weeks gestational age and the most common chronic lung disease in infancy.1 While survival of premature infants has improved significantly over the last few decades, despite numerous randomized controlled clinical trials, the incidence of BPD is unchanged. Animal models of BPD have greatly improved our understanding of the cellular and molecular underpinnings of BPD, but novel approaches that include human samples are needed to improve preventative and therapeutic strategies for BPD.2 Tracheal aspirates can be readily collected from intubated newborns and have been studied as a source of biomarkers for BPD.3,4 Mesenchymal stromal cells (MSCs) derived from tracheal aspirates have been studied by multiple groups, including by us.5,6 We have demonstrated that the transcriptome of lung MSCs correlates with degree of lung developmental maturity and BPD.6 However, epithelial cells derived from tracheal aspirates have not been studied. Recently, Wang et al. reported pediatric human lung epithelial cells derived from infant lung tissue, but the cells are heterogeneous, showed limited expansion ability, and were derived from autopsy samples.7 We have previously developed a culture system that allows for long-term expansion and subsequent differentiation of human epithelial basal cell-like cells.8,9,10 Here we show that epithelial basal cell-like cells can be expanded from newborn tracheal aspirates by modifying our existing protocol. We envision human tracheal aspirate-derived airway basal cell-like cells will be an important model system to study late-stage human lung development and perinatal lung disease.
Methods
Tracheal aspirate collection
Tracheal aspirates were collected during routine suctioning of intubated newborns. All the samples were collected using consents from protocols approved by the Institutional Review Board of Partners HealthCare.
Neonatal tracheal aspirate-derived basal cell-like cells (nTAD BCC) culture
Tracheal aspirates collected from intubated infants were stored at 4 °C for no more than 24 h prior to being cultured on plates precoated with laminin-enriched 804G-conditioned medium11,12 in small airway epithelial cell growth medium (PromoCell) with A-8301 (1 μM) and Y27632 (5 μM). Rapamycin, PP242, and Torin 1 were added at indicated concentrations. If the cultures are inspected every 1–2 days, epithelial cells colonies first become apparent by or before about 7 days as tight clusters of 5–20 cells. After these colonies appear, we allow them to expand an additional 7–10 days to have sufficient number of cells for the first passage (in part this depends on the number of colonies in the well). In some cases, there were small patches of nondividing large cells attached to the plate which were scraped off prior to passaging. For serial expansion of nTAD BCCs with or without rapamycin, at each passage, cells were split 1:8 when they reached confluency (equivalent to three population doublings per passage) until proliferation ceased.
Air−liquid interface (ALI) differentiation
Cells were trypsinized and seeded onto 0.4 μm Transwell membranes (3460, Corning) precoated with 804G-conditioned medium with a density of >6000 cells/mm2 (high epithelial cell seeding densities are critical for mucociliary differentiation) in medium composed of 50% growth medium (above), 25% Pneumacult-ALI medium (StemCell Technology, Cat. 05001), and 25% Dulbecco’s Modified Eagle’s Medium with 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid. (We found that this adaptation step (12–24 h) greatly reduces cell death.) The airway basal cell-like cells must be 100% confluent prior to differentiation. If the cells are not confluent following attachment, further expansion can be achieved using growth medium for a maximum of 2 days (both upper and lower chambers) until the cells reach confluence. Twelve hours after cell attachment/reaching confluency, excess cells were removed, and the medium was replaced with complete Pneumocult-ALI medium filling both the upper and lower chambers. The following day, the Pneumacult-ALI medium was added only in the lower chamber to create the airway−liquid interface. Early phase of cell stratification (formation of multiple layer from the initial monolayer of cells) is critical for high efficiency of differentiation; thus, for the first week of ALI culture, 5 μM ROCKi and 0.5 μM A-83-01 was included and the medium was changed daily until the differentiation was well established. Then, the differentiation was maintained for another 1–2 weeks with every other day medium change. Upon initiation of air−liquid interface, the cells undergo stratification and mucociliary differentiation, which is monitored daily under inverted-phase microscopy. Stratification was defined by a gradual decrease in transparency of membranes and increased crowding of the cells. Ciliogenesis was assessed by the appearance of ciliated cells. Based on our prior time course study, mucociliary differentiation plateaued after 11–14 days of ALI culture under our culture condition.8 Therefore, we routinely fix and stain ALI membrane for cell type markers after 14–18 days of ALI culture, but this may vary by cell line.
Immunofluorescence staining
Cells in culture or after differentiation on Transwell were fixed with 4% paraformaldehyde in phospate-buffered saline (PBS) for 15–30 min at room temperature. Some ALI membranes were further embedded in optimum cutting temperature compound for 7 μm frozen sections. For staining, cells or sections were first blocked and permeabilized with PBS + 3% bovine serum albumin (BSA) + 0.2% Triton X-100 for 1 h and then incubated with the primary antibodies diluted in PBS + 1% BSA at 4 °C overnight. Following incubation, the cells were rinsed three times with PBS + 0.2% Triton X-100 and incubated with Alexa Fluor-conjugated secondary antibodies (Life Technologies) at room temperature for 1–2 h. After rinsing three times with PBS + 0.2% Triton X-100, the nuclei were stained with 4′,6-diamidino-2-phenylindole. Samples were mounted or kept in PBS for imaging with a Nikon Ti inverted fluorescence microscope. Primary antibodies used were: NKX2.1 (sc-13040, Santa Cruz Biotechnology), p63 (GTX102425, GeneTex), CK5/6 (M7237, DAKO), EpCAM (12-9326-42, eBioscience), acetylated Tubulin (T7451, Sigma), MUC5AC (MA5-12178, Invitrogen), and CC10 (a.k.a CCSP or SCGB1A1, HPA031828, Sigma). Dilution was as follows for immunofluorescence: 1:200 for NKX2.1, p63 antibodies; 1:500 for CK5/6, MUC5AC, and acetylated tubulin antibodies; 1:100 for EpCAM and CC10 antibodies; 1:200 for secondary antibodies. Dilution was as follows for western blot: 1:500–1000 for primary antibodies; 1:2000 for secondary antibodies.
Western blotting
Cells were lysed with RIPA buffer with protease inhibitors and phosphatase inhibitors. Protein samples were separated through sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes, which were then blocked with 5% skim milk in tris(hydroxymethyl)aminomethane-buffered saline, 0.1% Tween 20 detergent (TBST) and incubated with primary antibodies overnight at 4 °C. Following washes with TBST, membranes were incubated with HRP-conjugated secondary antibodies for 1 h at room temperature and finally developed with ECL reagents. Volume tools of Bio-Rad Image Lab software were used to quantify band intensity. Primary antibodies were mTOR (2983, Cell Signaling Technology), phospho-mTOR (2974, Cell Signaling Technology), S6 (sc-74459, Santa Cruz Biotechnology), phospho-S6 (5364, Cell Signaling Technology), and β-actin (612656; BD Biosciences).
Seq-Well single-cell RNA-sequencing (scRNAseq)
Cells in fresh tracheal aspirates were washed and red blood cells removed using EasySep RBC depletion kit (18170, Stem Cell Technologies). Then 20,000 cells were loaded on a Seq-Well slide and processed to barcoded single-cell libraries as previously described.13 The barcoded libraries were sequenced on an Illumina NextSeq at Koch Institute core facility. Single-cell digital gene expression matrices were generated from the sequencing data as previously described.14 Mitochondrial reads were used as a filtering criterium as previously reported.13 Cells with fewer than 800 transcripts or 400 genes detected were excluded as low quality. The single-cell transcript counts were library-size normalized. Gene expression values for each cell were scaled by the total number of transcripts and then multiplied by the median transcript count of all cells. Finally, the values were log transformed and imported into Seurat for analysis.15 Expression data were reduced to 18 principal components that accounted for the majority of the variance in variable genes. Fifteen clusters of cells were identified in the data following further dimensionality reduction using t-SNE algorithm followed by density clustering (DBcust) in Seurat. Gene ontology analysis of cluster enriched genes using DAVID bioinformatics database. The marker genes of each cluster identified by Seurat were queried against human GTEx database using R/Bioconductor package TissueEnrich and were queried against LGEA sorted cells (Human Neo-to-Adult) data.16 RNA-seq data will be available through GEO under GSE137950.
Bulk level RNA-sequencing and analysis
For population-level RNA-seq, RNA samples from cells were extracted using Qiagen RNA RNeasy Kit and subjected to library preparation with Illumina TruSeq V2 kit. Sequencing was done at Mayo Clinic Next Generation Sequencing facility. Reads were aligned using STAR version 2.6.0a to the UCSC GRCh38 genome build and counts were calculated against the corresponding gene annotation using htseq-count version 0.10.0.17,18 Differential expression analysis was performed using R package DESeq2 1.10.1.19 Differentially expressed genes between paired rapamycin and no-rapamycin cells were determined based on absolute value of log2 fold change > 1 (twofold difference) in normalized counts. Gene set enrichment analysis (GSEA) was performed using the whole transcriptome ranked by fold changes according to Subramanian et al. against the gene sets described in MSigDB.20,21 Gene sets showing false discovery rate < 0.25 were deemed enriched and the normalized enrichment score which is the primary statistic that accounts for differences in size of gene sets to represent the scores for enriched gene sets were used to represent the scores of the enriched gene sets. Functional overrepresentation analysis of differentially expressed genes was performed using STRING v11.22 RNA-seq data are available through GEO under GSE137353.
Quantitative reverse transcription PCR
Total RNAs were reverse transcribed using Bio-Rad iScript reagent per manufacturer’s protocol. qPCRs were done with Bio-Rad EvaGreen SsoFast qPCR reagent and a Bio-Rad CFX96 Real-Time system in triplicates. Reference gene was GAPDH. Primers used were: RPS6KA2 Forward primer GAAAGCTCTACCTGATCCTGG; reverse primer GGCTGTGGAGATGGTCTAAAG; GAPDH forward primer ACATCGCTCAGACACCATG; reverse primer TGTAGTTGAGGTCAATGAAGGG.
Flow cytometry
For flow cytometry analysis of cell sizes, cells were fixed with 70% ethanol and analyzed with a BD Biosciences LSR cell analyzer. Cell size was analyzed based on forward scatter (FSC) distribution.
Trans-epithelial electrical resistance (TEER)
TEER was measured with the EVOM2, Epithelial Voltohmmeter (World Precision Instruments, Inc., Sarasota, FL).
Results
Newborn tracheal aspirates contain epithelial cells
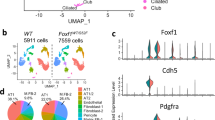
To investigate whether newborn tracheal aspirates contain epithelial cells, we performed single-cell RNA-sequencing (scRNAseq) on fresh aspirate samples collected from two infants (one sample each) born at 24–25 weeks gestational age (GA) using Seq-Well technology.13 Among the 867 cells identified in the two samples, we detected multiple types of immune and inflammatory cells, mesenchymal cells, and two clusters of epithelial cells based on established gene signatures, which include epithelial cell adhesion molecule (EpCAM) and cytokeratin (KRT or CK) (Fig. 1 and Supplementary Figs. S1 and S2). We analyzed the gene signatures of these two epithelial cell-clusters using the Lung Gene Expression Analysis (LGEA) Web Portal and the human Genotype-Tissue Expression (GTEx) database to confirm that they were derived from lung tissue (Supplementary Fig. S2 and Tables S1 and S2).23,24 However, when compared to lung airway scRNAseq data, these cells did not correlate with any specific lung epithelial cell sub-type (Supplementary Fig. S2).25,26 Nonetheless, our scRNAseq analysis demonstrated the presence of epithelial cells in newborn tracheal aspirates.
Selective culture of basal cell-like cells from tracheal aspirates
Basal cell-like cells in the epithelial population are likely rare and require further selection and expansion. We applied our recently published Rho/SMAD signaling inhibition protocol to tracheal aspirates, in order to selectively culture these progenitor cells by inhibiting their differentiation.8 When tracheal aspirates were seeded in this selection condition, tight monolayer epithelial cell colonies with typical epithelial cell morphology grew from plated samples within 2–7 days. These colonies were subsequently expanded to confluence in six-well plates and could be passaged 2–4 times at 1:3 or 1:4 ratio and survived freeze-thaw cycles. Approximately 40% of plated tracheal aspirate samples generated a cell line. Because intubated preterm infants are often suctioned multiple times per day, we were able to collect sufficient samples to establish cell lines from 80% of the subjects enrolled in our study. We refer to these cells as neonatal tracheal aspirate-derived (nTAD) epithelial basal cell-like cells (BCCs).
Although we were able to obtain basal cell-like cells from many infants, it became apparent that most of these cells deteriorated after a few passages. By passage 5 (sometimes as early as passage 2), the cells began to lose their epithelial phenotype. Instead of the typical cobblestone morphology, the cells became enlarged and elongated with multipolar processes (Fig. 2a and Supplementary Fig. S3). Moreover, their proliferation slowed and eventually ceased (Fig. 2b). These changes in proliferation and morphology after several passages were a feature of nearly all nTAD BCC lines, and only a few lines could be passaged beyond passage 5 without demonstrating obvious morphologic heterogeneity and limited capacity for expansion.
a Representative images of one nTAD BCC line showing enlarged, heterogenous cells without rapamycin and typical homogenous epithelial morphology with treatment. b Growth curve of three nTAD BCC lines showing decreased proliferation without rapamycin and after rapamycin withdrawal (black diamonds). Dotted line indicates BCC line proliferation ceased.
mTOR inhibition by rapamycin allows long-term expansion of nTAD BCC cells
The mammalian target of rapamycin (mTOR) plays a critical role in cell growth and its inhibitor rapamycin has been shown to prolong in vitro growth of epidermal and corneal epithelial cells and allow in vivo maintenance of tracheal basal cells.27,28,29 Therefore, we investigated whether addition of rapamycin could improve nTAD BCC cell culture. We introduced rapamycin to nTAD BCC cultures that had begun to demonstrate the aforementioned changes, but prior to significant deterioration (i.e. passages 2–4). With rapamycin, the cells appeared to go through an adaptation phase during which they became smaller and their proliferation slowed. 2–3 passages later, the cells had returned to their uniform, typical, cobblestone epithelial morphology (Fig. 2a and Supplementary Fig. S3). At rapamycin concentration of >5 nM, cell growth remained slow (not shown), but at 5 nM, cellular proliferation recovered towards exponential growth (Fig. 2b). We also tested other structurally unrelated mTOR inhibitors PP242 and Torin 1 and discovered that PP242 has a similar effect as rapamycin on cell morphology and proliferation, while Torin 1 had cell toxicity at low dose (Supplementary Fig. S3).30 Because PP242 and Torins have potentially stronger long-term effect on proliferation and rapamycin has been tested in other cellular systems, we focused our further experiments on rapamycin.27,28,29,31
Rapamycin-treated nTAD BCC lines could be maintained in culture for at least 15 passages (Fig. 2b). Continued mTOR inhibition was required—cell culture quickly deteriorated upon withdrawal of rapamycin (Fig. 2b). Addition of rapamycin upon initial plating of tracheal aspirates did not enhance BCC derivation (data not shown), suggesting that mTOR inhibition does not change the selective effect of the culture medium. We have established a repository of nTAD BCC lines from infants ranging in gestational age from 24 weeks to full term and with a variety of clinical conditions (Supplementary Table S3).
nTAD epithelial cells maintain progenitor cell properties in long-term culture
We examined the progenitor cell properties of nTAD BCCs. At low passage (2–4), all nTAD BCCs we tested, with or without rapamycin, stained positive for basal cell markers (p63, EpCAM and CK5), the lung endoderm transcription factor NKX2.1 (Fig. 3a), as well as other airway basal cell markers such as NGFR and PDPN (Supplementary Fig. S5). BCC lines cultured in rapamycin at later passages maintained consistent expression of these basal cell markers (Fig. 3b). Next, we tested the ability of nTAD BCCs to differentiate into functional airway epithelial cell types. Without rapamycin, only a few lines were able to be expanded sufficiently while maintaining healthy morphology to provide the number of cells required for air−liquid interface (ALI)-based differentiation. Figure 4 shows the differentiation potential of one nTAD BCC line derived with rapamycin from the initial tracheal aspirate plating. ALI differentiation of the early-passage cells generated a pseudostratified epithelium containing ciliated, goblet, and club cells in 2–3 weeks (Fig. 4a). With rapamycin, these cells retained an overall similar capacity to differentiate, even at passage 16 (Fig. 4b). However, when rapamycin was withdrawn from passage 3, the same cell line failed to form properly stratified epithelium at passage 8 and the differentiation was very inefficient and heterogenous (Fig. 4c). These data demonstrate that nTAD cells are bona fide epithelial BCs. Integrity of the ALI cultures was determined grossly by lack of culture media leaking into the apical side of the Transwell and via visual inspection on light microscopy. Cellular barrier integrity was further confirmed by measurements of trans-epithelial electrical resistance greater than 150 Ω cm2 (Supplementary Fig. S6). Rapamycin not only preserves the ability of the nTAD BCC lines to proliferate in an undifferentiated state, but also preserves their ability to give rise to differentiated progeny.
Rapamycin affects cellular proliferation and differentiation
mTOR and related pathways participate in the functions of pleiotropic cellular processes.32 We examined the mTOR signaling pathway and confirmed that rapamycin inhibited the phosphorylation of mTOR and its main target protein ribosomal S6 (Fig. 5), as expected. In order to understand the mechanisms mediating the effect of mTOR inhibition, we performed RNA-sequencing on two nTAD BCC lines (BCC11 and BCC13) that were each cultured with or without rapamycin (total of four samples) and examined the differentially expressed genes. Interestingly, GSEA identified clear enrichment of genes positively regulating cell proliferation for BCC11 and depletion of genes negatively regulating proliferation for BCC13, both promoting cell proliferation (Fig. 6a). In addition, there was depletion of genes involved in epithelial differentiation in both lines treated with rapamycin (Fig. 6b).
a GSEA revealed depletion of genes involved in epithelial differentiation in two nTAD BCC lines and overall positive regulation of epithelial proliferation. b Heatmap demonstrates genes involved in epithelial differentiation were downregulated with rapamycin treatment in two nTAD BCC lines (using differential gene expression analysis with twofold log change as cutoff). c RNA-seq data (left panel) shows higher RPS6KA2 level in two nTAD BCC lines. d Confirmation of higher RPS6KA2 levels using reverse transcription qPCR in a third line at passage 6 (NO RAPA: never cultured in rapamycin) and 11 (NO RAPA: rapamycin was maintained in culture until passage 9).
Next, we examined which mTOR pathway genes were specifically affected by rapamycin treatment. Among mTOR signaling pathway genes from Kyoto Encyclopedia of Genes and Genomes (KEGG), 127 were expressed in the nTAD BCCs.33 With mTOR inhibition, some of these canonical mTOR signaling pathway genes were downregulated in both lines. A few were upregulated with rapamycin and RPS6KA2 (a.k.a. RSK3) was the only one upregulated in both lines (Fig. 6c, d). The latter finding is particularly interesting, because RSK3 has been shown to support proliferation of breast cancer cells upon mTOR inhibition.34 Together these findings indicated that mTOR inhibition enhances long-term expansion of nTAD BCCs via two related mechanisms: promoting proliferation and inhibiting differentiation.
Discussion
Despite remarkable achievements over the past few decades, the prolonged expansion of adult epithelial basal cells remains challenging.35,36,37 Most primary human bronchial epithelial cells are generated from invasive procedures (e.g. bronchoscopy) or from autopsy specimen, thus limiting their clinical utility.7,8 These challenges significantly limit our ability to understand human lung development and disease in preterm infants. To overcome these critical limitations, we propose to use lung progenitor cells isolated from patient’s tracheal aspirates, which are readily collected during routine suctioning of intubated newborns. Here, we demonstrated that tracheal aspirates from intubated newborns contain epithelial cells and we applied Rho/SMAD/mTOR triple inhibition to enable long-term expansion of epithelial basal cell-like cells, capable of differentiating into pseudostratified airway epithelium.
Tracheal aspirates obtained from newborns (half milliliter to about 3 ml in volume) contain very few epithelial cells (<100). We have previously demonstrated that Rho/SMAD inhibition selectively allows the proliferation of BCs within a cell population.8 Using this method, we can reliably and consistently isolate BCCs from 80% of intubated infants. The initial outgrowth (p0) is usually from a few isolated colonies, each likely arising from individual cells. Hypothetically, from a tracheal aspirate that gives rise to four founder colonies each arising from a single cell, at least 18 population doublings would be required to generate 1 million cells—the amount needed for two 12-well Transwells—in our ALI differentiation protocol. Because these cells are not transformed, this initial period of proliferation likely limits the ability to passage much beyond cell line establishment. Indeed, in reported in vitro culture systems, common obstacles limiting their experimental utility are senescence and loss of functionality.7,8 Our discovery that adding mTOR inhibition extends the proliferative capacity of these cells is a significant improvement and allows enough material to perform important biological assays. nTAD BCCs can undergo ~50 population doublings during in vitro expansion. Since the basal cell-like cells expanded with Rho/SMAD inhibition do not express telomerase (our RNA-seq data), this phenomenon is likely consistent with the Hayflick limit or the maximum lifespan of human somatic cells without being immortalized.8,38
Transcriptomic analysis showed that mTOR inhibition help nTAD BCC expansion by promoting proliferation and suppressing differentiation of BCCs, in particular by upregulating RSK3. We did not find genes involved in oxidative stress, epithelial−mesenchymal transition, or cellular senescence to be differentially expressed with rapamycin treatment, as reported for some other cell types.27,29,39
We have confirmed that nTAD BCCs are lung derived. But we do not know the precise anatomical origin of the cells within the lung and it is possible that their ex vivo phenotype may depend on whether they arrived from proximal or more distal airway. Ultimately correlating our findings in nTAD BCC lines with samples for which the anatomic origin is more clearly delineated (e.g. surgical, bronchoscopy, or post-mortem) and applying single-cell analysis directly will answer this question. Cell types (e.g., mesenchymal and epithelial) identified using scRNA-seq could be compared to existing data in the LungMAP database and the forthcoming fetal lung single-cell RNA-seq data from Human Developmental Cell Atlas to refine our understanding of the developmental stage and anatomical origin of nTAD BCCs.16,40 We confirmed that nTAD BCCs expanded with rapamycin maintain their ability to differentiate to ciliated cells and secretory cells, even at late passages. There are many rare terminally differentiated cell types in the airway that we did not attempt to detect. Further experimentation is needed to examine the full differentiation potential of nTAD BCCs and how gestational age (developmental stage), clinical course, and potentially different anatomical origins affect their cell fate specification.
In a prior proof-of-concept experiment, we generated a comprehensive panel of neonatal tracheal aspirate-derived mesenchymal stromal cell lines (nTAD MSCs) derived from patients with gestational age spanning 23–42 weeks. We demonstrated that the transcriptional dynamics of nTAD MSCs correlate with lung developmental gene signatures and with molecular perturbations (e.g. abnormal extracellular matrix regulation) described in BPD. Together with the nTAD BCCs reported here, we envision a novel cellular model system to better understand BPD. BPD is a complex lung injury syndrome with prenatal origins that is characterized by alterations in normal lung development due to a myriad of postnatal insults, including oxygen, mechanical ventilation, and inflammation. As is the case with other chronic respiratory disorders including asthma and COPD, one major challenge is that the clinical definition of BPD fails to identify disease endotypes—subsets of patients with distinct pathobiological mechanisms. This concept explains the clinical heterogeneity of BPD and the low success rate of clinical trials—often a therapeutic intervention is more likely to target one endotype and therefore will be ineffective for patients with the same disease, but with a different endotype. Additionally, the lung and its function are difficult to access (especially in newborns) and thus we must rely on biomarkers. Tracheal aspirate-derived lung progenitor cells could serve as a biomarker to help stratify BPD into endotypes, but this will require rigorously characterizing and phenotyping the function of nTAD progenitor MSCs and BCCs. The nTAD BCC biorepository that we are developing using our Rho/SMAD/mTOR inhibition protocol described will be instrumental in these future efforts (Supplementary Table S3).
References
Jobe, A. H. Mechanisms of lung injury and bronchopulmonary dysplasia. Am. J. Perinatol. 33, 1076–1078 (2016).
McEvoy, C. T. et al. Bronchopulmonary dysplasia: NHLBI Workshop on the Primary Prevention of Chronic Lung Diseases. Ann. Am. Thorac. Soc. 11(Suppl 3), S146–S153 (2014).
Kotecha, S., Wangoo, A., Silverman, M. & Shaw, R. J. Increase in the concentration of transforming growth factor beta-1 in bronchoalveolar lavage fluid before development of chronic lung disease of prematurity. J. Pediatr. 128, 464–469 (1996).
Thompson, A. & Bhandari, V. Pulmonary biomarkers of bronchopulmonary dysplasia. Biomark. Insights 3, 361–373 (2008).
Hennrick, K. T. et al. Lung cells from neonates show a mesenchymal stem cell phenotype. Am. J. Respir. Crit. Care Med. 175, 1158–1164 (2007).
Spadafora, R. et al. Lung-resident mesenchymal stromal cells reveal transcriptional dynamics of lung development in preterm infants. Am. J. Respir. Crit. Care Med. 198, 961–964 (2018).
Wang, Q. et al. A novel in vitro model of primary human pediatric lung epithelial cells. Pediatr. Res. 87, 551–517 (2019).
Mou, H. et al. Dual SMAD signaling inhibition enables long-term expansion of diverse epithelial basal cells. Cell Stem Cell 19, 217–231 (2016).
Peters-Hall, J. R. et al. Long-term culture and cloning of primary human bronchial basal cells that maintain multipotent differentiation capacity and CFTR channel function. Am. J. Physiol. Lung Cell Mol. Physiol. 315, L313–L327 (2018).
Zhang, C. et al. Long-term in vitro expansion of epithelial stem cells enabled by pharmacological inhibition of PAK1-ROCK-Myosin II and TGF-beta signaling. Cell Rep. 25, 598–610 e595 (2018).
Langhofer, M., Hopkinson, S. B. & Jones, J. C. The matrix secreted by 804G cells contains laminin-related components that participate in hemidesmosome assembly in vitro. J. Cell Sci. 105(Pt 3), 753–764 (1993).
Levardon, H., Yonker, L. M., Hurley B. P. & Mou, H. Expansion of airway basal cells and generation of polarized epithelium. Bio. Protoc. 8, pii e2877 (2018).
Gierahn, T. M. et al. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods 14, 395–398 (2017).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Du, Y. et al. Lung Gene Expression Analysis (LGEA): an integrative web portal for comprehensive gene expression data analysis in lung development. Thorax 72, 481–484 (2017).
Anders, S., Pyl, P. T. & Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Szklarczyk, D. et al. STRING v11: protein−protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Consortium, G. T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Jain, A. & Tuteja, G. TissueEnrich: tissue-specific gene enrichment analysis. Bioinformatics 35, 1966–1967 (2019).
Montoro, D. T. et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324 (2018).
Plasschaert, L. W. et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377–381 (2018).
Gidfar, S. et al. Rapamycin prolongs the survival of corneal epithelial cells in culture. Sci. Rep. 7, 40308 (2017).
Haller, S. et al. mTORC1 activation during repeated regeneration impairs somatic stem cell maintenance. Cell Stem Cell 21, 806–818 e805 (2017).
Iglesias-Bartolome, R. et al. mTOR inhibition prevents epithelial stem cell senescence and protects from radiation-induced mucositis. Cell Stem Cell 11, 401–414 (2012).
Feldman, M. E. et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 7, e38 (2009).
Guertin, D. A. & Sabatini, D. M. The pharmacology of mTOR inhibition. Sci. Signal. 2, pe24 (2009).
Saxton, R. A. & Sabatini, D. M. mTOR signaling in growth, metabolism, and disease. Cell 169, 361–371 (2017).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Serra, V. et al. RSK3/4 mediate resistance to PI3K pathway inhibitors in breast cancer. J. Clin. Invest. 123, 2551–2563 (2013).
Liu, X. et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 180, 599–607 (2012).
Rheinwald, J. G. & Green, H. Formation of a keratinizing epithelium in culture by a cloned cell line derived from a teratoma. Cell 6, 317–330 (1975).
Wang, X. et al. Cloning and variation of ground state intestinal stem cells. Nature 522, 173–178 (2015).
Hayflick, L. & Moorhead, P. S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 25, 585–621 (1961).
Xiang, S. et al. Rapamycin inhibits epithelial-to-mesenchymal transition of peritoneal mesothelium cells through regulation of Rho GTPases. FEBS J. 283, 2309–2325 (2016).
Behjati, S., Lindsay, S., Teichmann, S. A. & Haniffa, M. Mapping human development at single-cell resolution. Development 145, pii: dev152561 (2018).
Acknowledgements
This work was supported by the Department of Pediatrics, Massachusetts General Hospital. This work was also partly supported by the Harvard Stem Cell Institute (H.M.) and the Charles H. Hood Foundation (H.M.).
Author information
Authors and Affiliations
Contributions
J.L.: Substantial contributions to conception and design, acquisition of data, and analysis and interpretation of data; drafting the article or revising it critically for important intellectual content. X.Z.: Substantial contributions to conception and design, acquisition of data, and analysis and interpretation of data. J.E.S., L.X., T.G., C.Z., M.W., B.C.C.: Acquisition of data, and analysis and interpretation of data. J.C.L., H.L.: Analysis and interpretation of data. S.H.: Acquisition of data. H.M.: Substantial contributions to conception and design, analysis and interpretation of data; revising the article critically for important intellectual content. P.H.L.: Substantial contributions to conception and design, analysis and interpretation of data; revising the article critically for important intellectual content, and final approval of the version to be published.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Patient consent
Patient samples were collected using informed consent forms approved by the Institutional Review Board of Partners HealthCare.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Lu, J., Zhu, X., Shui, J.E. et al. Rho/SMAD/mTOR triple inhibition enables long-term expansion of human neonatal tracheal aspirate-derived airway basal cell-like cells. Pediatr Res 89, 502–509 (2021). https://doi.org/10.1038/s41390-020-0925-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-020-0925-3
This article is cited by
-
Perinatal origins of bronchopulmonary dysplasia—deciphering normal and impaired lung development cell by cell
Molecular and Cellular Pediatrics (2023)
-
CircRNA, lncRNA, and mRNA profiles of umbilical cord blood exosomes from preterm newborns showing bronchopulmonary dysplasia
European Journal of Pediatrics (2022)
-
Long-term differentiating primary human airway epithelial cell cultures: how far are we?
Cell Communication and Signaling (2021)
-
Prematurity alters the progenitor cell program of the upper respiratory tract of neonates
Scientific Reports (2021)