Abstract
Background
Animal studies suggest that total parenteral nutrition (TPN) may alter bacterial colonization of the intestinal tract and contribute to complications. Progressive changes in gut microbiome of infants receiving TPN are not well understood.
Methods
Infants with and without TPN/soy lipid were enrolled in a prospective, longitudinal study. Weekly fecal samples were obtained for the first 4 weeks of life. High throughput pyrosequencing of 16S rDNA was used for compositional analysis of the gut microbiome.
Results
47 infants were eligible for analyses, 25 infants received TPN, and 22 infants did not (control). Although similar between TPN and control groups in the first week, fecal bacterial alpha diversity was significantly lower in the TPN group compared to controls at week 4 (Shannon index 1.0 vs 1.5, P-value = 0.03). The TPN group had significantly lower Bacteroidetes and higher Verrucomicrobia abundance compared to controls (P-values < 0.05), and these differences became more pronounced over time. At the genus level, TPN was associated with lower abundance of Bacteroides and Bifidobacterium in all weeks.
Conclusions
TPN is associated with significant loss of biodiversity and alterations in the pattern of gut microbial colonization of infants over time. TPN-associated dysbiosis may predispose infants to adverse NICU outcomes.
Similar content being viewed by others
Introduction
Total parenteral nutrition (TPN) is essential for the survival of critically ill infants who cannot be enterally fed. Early initiation of TPN in infants cared for in the neonatal intensive care unit (NICU) also improves brain growth and mediates the effects of critical illness on later growth and long-term neurodevelopmental outcomes.1,2 Despite these benefits, animal studies suggest that TPN may significantly alter the bacterial colonization of the intestinal tract.3 Lower overall microbial diversity and expansion of potentially pathogenic bacteria are seen in the intestinal microbiome of enterally deprived animals receiving TPN.4,5 Importantly, TPN-associated alterations in the gut microbiome is associated with increased expression of proinflammatory cytokines within the intestinal mucosa and loss of intestinal barrier function.4 To date, there are limited data regarding the longitudinal changes in the gut microbiome of humans receiving TPN.
Adverse outcomes in NICU infants such as necrotizing enterocolitis (NEC) and parenteral nutrition-associated liver disease (PNALD) have been associated with specific changes in the microbiome. NEC is a potentially life-threatening intestinal disease that occurs in approximately 7% of infants with birth weight less than 1500 gm.6 Death occurs in 15–42% of affected infants, and survivors are at risk for adverse long-term outcomes including neurodevelopmental delay.7,8 PNALD can be a devastating hepatic complication of long-term intravenous nutrition. The incidence of PNALD ranges from 40–85% in infants receiving prolonged TPN,9 with a clinical spectrum that ranges from steatosis and cholestasis to hepatic fibrosis, cirrhosis, and potential death.10 While the cause of both NEC and PNALD is likely multifactorial, alterations in the gut microbiome characterized by lower diversity, lower abundance of Bacteroidetes and Firmicutes, and higher abundance of Proteobacteria has been reported in infants with each disease, relative to controls.11,12
If TPN alters the microbiome, it may contribute to adverse outcomes in NICU infants. However, the progressive changes that occur in the gut microbiome of neonates receiving TPN are unknown. The primary objective of this study was to compare the fecal microbiome of infants receiving TPN and control infants who did not receive TPN. The secondary objective was to explore the impact of TPN while adjusting for other factors known to alter the microbiome of infants in the NICU.
Methods
Study subjects
This prospective, longitudinal cohort study was approved by the Institutional Review Boards at the Children’s Hospital of Wisconsin and Medical College of Wisconsin and conducted in the Children’s Hospital of Wisconsin Neonatal Intensive Care Unit (NICU) and the Froedtert Hospital Newborn Nursery between May 2014 and May 2016. Written informed consent was obtained from parents or legal guardians. Infants >32 weeks gestational age at birth were recruited for study participation if they did not have primary liver disease, anatomic liver anomalies, metabolic disease, or known genetic or chromosomal defects. This gestational age cutoff was chosen because of the higher likelihood of having infants of similar gestational age who did not receive TPN to serve as controls. Cases were infants who received TPN with soy lipid, controls were infants who did not receive TPN or soy lipid. Due to the potential for prolonged antibiotics to significantly influence the microbiota, infants who received antibiotics for >10 days were excluded from the study.
All participants received routine care as determined by the providing physician. The provision of TPN with soy lipid followed pre-established NICU guidelines. In general, intravenous amino acid (provided as TrophAmine, B. Braun, Irvine, CA) was initiated at a dosage of 3 gm/kg/day and advanced to a maximum of 3.5–4 gm/kg/day in preterm infants and a maximum of 3 gm/kg/day in term infants, soy lipid (provided as Intralipid 20%, Fresenius Kabi, Uppsala, Sweden) was initiated at 2 gm/kg/day and advanced to a maximum of 3 gm/kg/day, and carbohydrates were initiated at a glucose infusion rate of 4–6 mg/kg/minute and advanced to a maximum of 10–12 mg/kg/minute. Breast milk was the primary source of enteral nutrition if available, otherwise formula was provided. No probiotics were given to any study participant.
Demographic and clinical data were collected from the medical record. Demographic information included gestational age and growth parameters at birth, sex, race, and mode of delivery. Clinical data consisted of diagnoses and surgical procedures. Daily nutritional, medication, and laboratory data including total and conjugated bilirubin were recorded. Cholestasis was defined as a serum conjugated bilirubin ≥ 2 mg/dL. Study data were collected and managed using the Research Electronic Data Capture (REDCap) system, a secure, web-based application used for data capture and management in clinical and translational research.13
Sample collection and processing
Fecal samples were obtained weekly for the first 4 weeks of life. If more than one stool was collected for a given week, only the first stool obtained in the week was used. For each collection, fresh stool was placed in RNAlater, rocked overnight at 4 °C, and stored at −80 °C until processed. For infants discharged home before a month of age, parents/guardians were provided with collection kits, including sterile vials prefilled with RNAlater, and instructed on the proper procedures for stool collection. Fecal samples collected by parents/guardians were returned via overnight mail for the remaining weeks of stool collection, and were rocked and stored in a similar fashion until processed.
Microbial genomic DNA was isolated from fecal samples using the MoBio PowerSoil® DNA Isolation kit (MoBio, Carlsbad, CA) following the manufacturer’s guidelines. Polymerase chain reaction (PCR) amplification using primers targeting the V4 region of the bacterial 16S rRNA gene, gene sequencing using the MiSeq platform (Illumina, San Diego, CA), and compositional analysis using the UPARSE algorithm and SILVA Database14,15 were performed by Diversigen, Inc. (Houston, TX). For a full description of PCR amplification gene sequencing, and compositional analysis, see Supplementary Methods.
Data analyses
Samples with low sequencing reads of less than 1000 were removed prior to data analyses. Identification of bacterial taxa and their abundance, and comparative analyses of samples followed protocols from the Human Microbiome Project (HMP).16,17 Briefly, bacterial taxonomy (genus and species) was quantified by determining operational taxonomic units (OTUs) and richness of the microbiota was determined using the total number of taxonomic units. The Shannon diversity index, which represents the sum of the relative proportion of bacterial species, was used to determine microbial diversity within each sample (alpha diversity). A Bray–Curtis distance matrix of normalized OTU counts and principal coordinate analysis were used to compare the distribution of bacterial species among samples (beta diversity). Permutational Multivariate Analysis of Variance (PERMANOVA) using distance matrices was used to compare beta diversity between groups; and a homogeneity of dispersion test was used to investigate the differences in group homogeneities. Bacterial diversity, presence or absence of specific bacterial groups, and quantitative differences in abundance were determined for each weekly stool sample of subjects receiving TPN and controls without TPN. Microbial abundance was standardized to the median sequencing depth for each sample. Sparse phyla with zero OTU in more than 75% of the data were excluded from the analyses because they were unlikely to contribute to the overall composition of any given sample.
Categorical data were compared using Chi-square or Fisher’s exact test. For continuous variables, normally distributed data were compared using t-test or ANOVA, and data that were not normally distributed were compared using a nonparametric Wilcoxon–Mann–Whitney test. All tests were conducted as two-tailed tests with a P-value < 0.05 considered statistically significant. Unless otherwise stated, the data are expressed as median and interquartile range (IQR).
A linear mixed model (LMM) was used to examine the relationship between TPN (yes vs no) and Shannon alpha diversity over time. An unstructured covariance structure was used to account for correlations within a patient. A generalized linear mixed model (GLMM) was used to determine the association between TPN and relative abundance over time. Proportions were transformed using an arcsine square-root transformation or logit transformation to ensure fit. Overall relative abundance of the different phyla by groups was summarized by means and the p-value was based on the model. For count data, to correct for overdispersion, a negative binomial distribution was used. The intercept was modeled as random. Maximum likelihood estimation was used and missing data were assumed to be at random. Stata Version 14.0 (Stata Corporation, College Station, TX), R (https://www.R-project.org/), and SAS Version 9.4 (SAS Institute, Cary NC) were used for statistical analyses.
Results
Study population
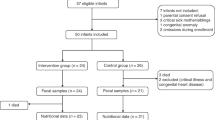
A total of 47 infants were eligible for analyses, 25 infants received TPN and 22 control infants did not receive TPN. Overall, study participants had a median gestational age of 36.1 weeks (IQR 34.9–37) and birth weight of 2.56 kg (IQR 2.27–3.03). Demographic and clinical data of infants who received TPN and controls are shown in Table 1. Gestational age and birth weight were similar between infants who received TPN and controls. There were also no differences in the distribution of gender, race, or mode of delivery between the two groups. Infants who received TPN were more likely to have a gastrointestinal (GI) diagnosis on admission, undergo GI surgery, and receive antibiotics. For infants who received TPN, median age at the start of TPN was 1 day of life (IQR 1–1) and median TPN duration was 20 days (IQR 14–25). Initiation of enteral feeds was significantly later in infants who received TPN. No patients received probiotics.
Overall taxonomic composition of the gut microbiota
A total of 134 fecal samples were available for analyses, with a median of three samples per patient in each group. An average of 22,712 gene sequences (range 3150–33,307) were generated per sample, and an average of 24 discrete bacterial taxa (or OTUs) were present per sample. Overall, the taxonomic composition of the gut microbiome of study infants was dominated by only a few phyla during the first month of life. Among all taxa, Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria, and Verrucomicrobia were the dominant phyla, representing 46%, 45%, 4%, 4%, and 1% of the total reads, respectively. Gammaproteobacteria, Bacilli, and Clostridia were the dominant classes (45%, 28%, and 11% of total reads, respectively).
Compositional differences in the microbiota: TPN versus Controls
Figure 1a shows group comparisons in the overall relative abundance of the different phyla between infants who received TPN and control infants who did not receive TPN. While there were subtle differences in the relative abundance of most phyla between groups, the proportion of Bacteroidetes was significantly higher in the control group compared to the TPN group (9% vs 0.1%, P-value < 0.001).
Taxonomic differences between TPN and control groups. a Pie chart showing the overall differences in the relative abundance of the most abundant phyla between TPN and control groups. Taxa with <1% relative abundance were combined as “Other”. b Stacked bar graphs showing longitudinal differences in the relative abundance of the dominant phyla between TPN and control groups in the first 4 weeks of life
Longitudinal differences in the composition of the gut microbiome between groups in the first 4 weeks of life are shown in Fig. 1b. In both groups, there was variability in the relative abundance of the different phyla from week to week. This variability appeared less pronounced for Firmicutes and Proteobacteria in weeks 3 and 4 in the control group. Bacteroidetes was again enriched in the control group, with a significant difference in its relative abundance in weeks 1, 2, and 4 compared to the TPN group (P-values < 0.05 for all comparisons). Group differences were also seen at the genus level. Differences in the relative abundance of the most dominant genera in each phylum between TPN and control groups in the first 4 weeks of life are shown in Fig. 2. In almost all weeks, infants in the TPN group had virtually no bacteria in the genera Bifidobacterium and Bacteroides, whereas infants in the control group had a paucity of bacteria in the genera Akkermansia compared to TPN infants. The top 10 most abundant genera in each group are shown in Supplemental Table 1.
Differences in biodiversity over time
Community-level comparisons of the gut microbiome between groups were also performed. Alpha diversity, or the richness and evenness of the species in the microbiome community, in each group is shown in Fig. 3. There was no difference in alpha diversity between groups in the first week of life. In both groups, diversity fell between the first and second weeks of life. For infants who received TPN, this lower diversity persisted through to week 4. In contrast, the diversity of control infants increased at the end of the month and was significantly higher than the TPN group (P-value = 0.03). In order to examine whether the relationship between TPN and alpha diversity over time was influenced by other factors, TPN and Shannon index were modeled with the inclusion of mode of delivery, treatment with proton pump inhibitor (PPI) or histamine-2 receptor antagonist (H2 blocker), and type of enteral feeding. Due to the small sample size, each covariate was assessed in the model with TPN exposure individually. Notably, the difference in alpha diversity between groups at week 4 remained significant when mode of delivery, treatment with PPI or H2 blocker, and type of enteral feeding were each included in the analyses (P-value < 0.05 for all comparisons). Beta diversity, or the microbial community compositional differences among samples, was explored using principal coordinates analysis (PCoA) on Bray–Curtis distance measurements in each group (Supplemental Figure S1). There were no differences in beta diversity between TPN and control groups for any week (P-values > 0.1 for all weeks), although the small sample size in this study may have precluded the detection of significant differences in overall community structure.
Community-level changes over time in TPN versus control groups. Differences in alpha diversity, or the richness and evenness of the species in the microbiome community, between TPN and control infants during the first 4 weeks of life. Shannon diversity index is shown on the y-axis and individual weeks are shown on the x-axis. Orange boxes indicate TPN group and blue boxes indicate control group. Box represents the 25th and 75th percentiles; line inside the box represents the median; error bars represent the minimum and maximum. *P-value < 0.05
Association of TPN exposure and differences in phyla abundance over time
Model estimates and standard error of phylum abundance in each group during the first 4 weeks of life are shown in Fig. 4. In almost all weeks, there was significantly lower abundance of Bacteroidetes and higher abundance of Verrucomicrobia in the TPN group compared to controls (P-value < 0.05 for all comparisons). These differences became more pronounced over time. The patterns of abundance over time and differences between TPN and control groups were relatively unchanged when mode of delivery and treatment with PPI or H2 blocker medication were each included in the model. (Supplemental Figure S2).
We explored the influence of enteral feeding on the compositional changes associated with TPN. Modeled estimates of phylum abundance were examined in weeks 1 and 4 in a subgroup of 10 TPN infants who received TPN in the first week of life and were weaned off of TPN and receiving full enteral feeds at week 4, compared to 10 control infants who also had fecal samples in both weeks 1 and 4 (Supplemental Figure S3). Despite the subsequent receipt of full enteral feeding, significantly lower Bacteroidetes and higher Verrucomicrobia was still observed in the TPN subgroup at the end of the study period.
Taxonomic composition of the gut microbiota of patients with cholestasis
Five infants in the TPN group were later found to have cholestasis. Median time to develop cholestasis was 29 days (IQR 24–29). Several of these infants had rare fecal samples in the first 4 weeks of life, making it difficult to assess progressive changes in the microbiome during this period. However, fecal samples were obtained and analyzed for each infant at the time they developed cholestasis (Fig. 5). In all five infants, Proteobacteria was the most prominent phylum, with a relative abundance of >69%.
Association of antibiotics and alpha diversity
The majority of infants in the TPN group received antibiotics, and antibiotic use in this group was complex with differences in types of intravenous antibiotics, dosages, durations of usage, and number of antibiotic courses among infants. While it was not possible to control for these differences, we sought to explore the impact of antibiotics and TPN on changes in microbial diversity of study infants over time. To do this, we compared the alpha diversity in the first 4 weeks of life of 10 TPN infants and 13 control infants who all received a short course of intravenous antibiotics for <3 days immediately after birth (Supplemental Figure S4). Although there was a trend toward lower alpha diversity in the TPN compared to control group at week 4, the difference was not significant (P-value = 0.22).
Discussion
This is the first study to characterize progressive changes in the microbiome of newborn infants receiving TPN. We identified differences in the gut microbiome of infants who receive TPN and those who did not. Although biodiversity is initially similar between groups after birth, TPN is associated with persistently low alpha diversity in the first 4 weeks of life. Infants receiving TPN have significantly lower abundance of Bacteroidetes and higher abundance of Verrucomicrobia compared to controls, and these differences between groups become more pronounced over time. Taken together, these findings suggest that enteral deprivation with TPN may alter the pattern of gut microbial colonization of NICU infants.
Laboratory studies using different animal models have reported significant changes in the microbiome associated with TPN-dependence and enteral deprivation. These changes are characterized by reduced bacterial diversity with a relative expansion of pathogenic bacteria.3 Compared to enterally fed newborn piglets, those receiving TPN had lower microbial diversity and enrichment of toxin-expressing or mucolytic Clostridium species.5,18 In mice, TPN administration was associated with a shift in the microbial composition from predominantly Firmicutes to predominantly Proteobacteria and Bacteroides.4
Data regarding changes in the microbiome of humans receiving TPN have been limited, with most prior reports involving older populations with specific diseases or conventional culture techniques. In children and adults with short bowel syndrome, those with TPN-dependence were reported to have lower intestinal microbial diversity and higher relative abundance of Proteobacteria compared to those who had been weaned from TPN and healthy controls.19,20 Adults with Crohn’s disease exhibited lower diversity and a significant increase in Enterococcus species after initiation of TPN.21 In a post hoc analysis of data obtained using traditional culture techniques and collected in a prospective study of preterm infants at risk for sepsis, Parm et al. found an association between TPN and reduced colonization of Gram-negative and Gram-positive bacteria.22 Our data further extend these observations to identify specific changes that occur in the developing microbiome over time in infants receiving TPN.
Consistent with the published literature, we found alterations in the gut microbiome in infants receiving TPN compared to control infants who did not receive TPN. We observed persistently low bacterial diversity in the TPN group over time. In both groups, there was a decline in alpha diversity by the second week of life. The etiology for this decline is unclear but may be associated with antibiotic exposure in many infants after birth. By the end of the study period, we found increased diversity in the control group. This diversification may represent processes involved in the maturation of the infant microbiome after birth. In a recent study of term infants, Chu et al. reported maturation of the newborn microbiome during the first several weeks of life with significant reorganization of microbial community structure characterized by expansion and diversification by the sixth week of life.23 In contrast, infants receiving TPN demonstrated persistently low diversity over the study period. These results suggest that, over time, TPN alters the colonization pattern and delays the maturation of the developing gut microbiome.
By analyzing serial fecal samples, we were able to model longitudinal changes in the microbiome to examine the pattern of microbial colonization in the two study groups. In all weeks, we found fewer Bacteroidetes, specifically Bacteroides, in infants receiving TPN compared to controls. Interestingly, Bifidobacterium were also lower in the TPN group throughout the study period. Lower abundance of these bacteria may have consequences for early intestinal immunity and resistance to colonization by pathogens. As putative beneficial commensals and pioneer bacteria in the newborn gut, both Bacteroides and Bifidobacterium are essential for colonization of the GI tract and educating the developing immune system.24 Capsular polysaccharide A of Bacteroides fragilis activates IL-10, and decreases activation of the innate immune response by increasing secretory IgA production and regulating the balance between helper T-cells.25,26 In vitro, secreted products from Bifidobacteria downregulate proinflammatory IL-8 and IFNγ cytokines and upregulate anti-inflammatory pathways by increasing IL-10 production.27 In addition, production of acetate by Bifidobacteria protects against colonization of pathogens.28 Given the role of Bacteroides and Bifidobacterium in immunotolerance and colonization resistance, it is possible that the persistently low abundance of these bacteria may predispose infants receiving TPN to intestinal inflammation and facilitate expansion of pathogenic bacteria.
Verrucomicrobia were significantly enriched in the infants receiving TPN relative to controls. Elevated Verrucomicrobia, particularly Akkermansia muciniphila, was similarly reported in mice following TPN.4 Akkermansia muciniphila is a mucous-degrading bacteria whose growth is favored when there is low availability of enteral nutrients such as during fasting and malnutrition.29 We suspect that the persistence of Verrucomicrobia in our TPN group is likely due to the lack of enteral nutrition that accompanies the TPN-dependent state, as we also observed the decline of this phylum in control infants who were enterally fed.
Interestingly, recent data suggest a link between Akkermansia muciniphilia and the intestinal mucosal immune system. Mice treated with Akkermansia muciniphilia had beneficial increases in intestinal regulatory T-cells and goblet cells.30 However, Seregin et al. found evidence that Akkermansia muciniphilia can act as a harmful pathobiont to promote colitis by upregulating proinflammatory cytokines in genetically susceptible mice.31 Whether Akkermansia is beneficial or harmful likely depends on genetic as well as environmental factors. Our finding of Akkermansia only in the TPN group is likely due to its ability to thrive during periods of nutrient deprivation; however, we postulate that persistent degradation of mucin by Akkermansia muciniphilia during prolonged periods of enteral deprivation may lead to a thinner intestinal mucous layer, loss of intestinal barrier function, and subsequent microbial-driven inflammation that has been reported with prolonged TPN administration.4
Although we found a trend toward higher abundance of Proteobacteria in infants receiving TPN over time, this did not reach statistical significance. Gram-negative bacteria, such as Proteobacteria, modulate the host’s immune system by promoting a proinflammatory state. In enterally deprived mice receiving TPN, an increase in the abundance of Proteobacteria was associated with increased toll-like-receptor (TLR) signaling and increased expression of proinflammatory cytokines within the intestinal lamina propria. These changes were accompanied by phenotypic signs of compromised intestinal barrier function including increased bowel wall permeability and mucosal atrophy.4 It has been hypothesized that the enteral deprivation in the TPN-dependent state promotes the selective growth of starvation-tolerant Proteobacteria.32 In this study, enteral feeds were initiated after a relatively short period of ~2 weeks in infants receiving TPN, and it is possible that longer periods of enteral deprivation may further promote Proteobacteria survival. Based on our findings, we also speculate that the lower abundance of beneficial commensals such as Bifidobacterium and Bacteroides may contribute to gradual enrichment of Proteobacteria in infants receiving TPN.
Mode of delivery, use of PPI or H2 blocker medication, and enteral feeding have all been reported to significantly alter the microbiome.33,34 Due to the small sample size of this study, we could not simultaneously control for all of these factors. However, when each was separately included into the analyses along with TPN, differences in alpha diversity and patterns of abundance of the dominant phyla between infants with TPN and controls remained significant, and indicates that TPN is an important factor affecting the developing gut microbiome. Larger studies are needed to further examine the combined effects of these variables.
The relevance of lower diversity and decreased relative abundance of Bacteroidetes in infants receiving TPN should be considered in the context of the alterations in the gut microbiome seen in diseases that affect NICU infants, such as NEC and PNALD. For both diseases, lower alpha diversity has been reported in affected individuals compared to controls.12,35 In a recent systemic review and meta-analysis of 14 studies comparing the intestinal microbiome of preterm infants who developed NEC to controls, Pammi et al. found that the microbiome was characterized by decreased relative abundance of Firmicutes and Bacteroidetes and increased relative abundance of Proteobacteria prior to development of NEC.11 To our knowledge, there have been no published microbiome studies performed in infants with NEC that specifically examined the association with TPN. However, provision of TPN is directly related to enteral deprivation, and feeding practices that prolong enteral deprivation may increase the risk for NEC.36,37
Infants with short bowel syndrome and PNALD have been reported to have lower overall diversity and a shift from Firmicutes to Proteobacteria.38 In five infants who developed PNALD, we found that all had a predominance of Proteobacteria at the time of cholestasis. The lack of serial fecal samples in these infants preclude our ability to determine progressive changes in the microbiome that may have predisposed to development of the disease. Future studies with longer time periods are needed to better understand the alterations in the gut microbiome that occur with prolonged TPN use.
This study has limitations. The small size and short time course of our study limits our ability to examine how the microbiome changes with more prolonged TPN and the association of these changes with clinical outcomes. The majority of infants in our TPN group had enteral deprivation and GI pathology. While it is difficult to separate the influence of these factors on our findings, critically ill infants seldom undergo enteral deprivation without TPN in clinical practice. Current data in animals and humans suggest that the lack of enteral nutrition, as opposed to TPN itself, underlies the intestinal phenotype seen in the TPN-dependent state.39,40 The clinical care of infants in the NICU is often complex, and it’s uncertain how other NICU practices may contribute to the changes we observed. Only soy-based lipids were provided to infants receiving TPN, and the impact of alternative lipid emulsions on the infant gut microbiome require further investigation.
While 16S rDNA allowed us to efficiently examine the bacterial composition of the gut microbiome at multiple time points, it is a targeted approach and provides little information regarding microbial function. Multiple “-omic” approaches will be needed to further understand microbial-host interactions. These include metagenomic approaches that examine the entire gene content of communities to capture the functional potential of a microbial community; transcriptomics to identify active functional profiles of a microbial community under specific conditions; and metabolomics for information about interactions within a microbial community and between the microbial community and the host environment.
Conclusions
In this longitudinal cohort study, we found significant differences in the microbiome of NICU infants who received TPN and control infants who did not receive TPN. TPN is associated with persistently low alpha diversity in the first 4 weeks of life. Over time, there is lower diversity as well as a relative loss of Bacteroidetes and enrichment of Verrucomicrobia that occurs in the gut microbiome of NICU infants receiving TPN compared to controls. We speculate that TPN-associated alterations in the gut microbiome may predispose NICU infants to adverse outcomes such as NEC and PNALD.
References
Tan, M., Abernethy, L. & Cooke, R. Improving head growth in preterm infants—a randomised controlled trial II: MRI and developmental outcomes in the first year. Arch Dis. Child Fetal Neonatal Ed. 93, F342–F346 (2008).
Ehrenkranz, R. A. et al. Early nutrition mediates the influence of severity of illness on extremely LBW infants. Pedia. Res. 69, 522–529 (2011).
Demehri, F. R., Barrett, M. & Teitelbaum, D. H. Changes to the intestinal microbiome with parenteral nutrition. Nutr. Clin. Pr. 30, 798–806 (2015).
Miyasaka, E. A. et al. Total parenteral nutrition-associated lamina propria inflammation in mice is mediated by a MyD88-dependent mechanism. J. Immunol. 190, 6607–6615 (2013).
Deplancke, B. et al. Selective growth of mucolytic bacteria including Clostridium perfringens in a neonatal piglet model of total parenteral nutrition. Am. J. Clin. Nutr. 76, 1117–1125 (2002).
Fanaroff, A. A. et al. Trends in neonatal morbidity and mortality for very low birthweight infants. Am. J. Obstet. Gynecol. 196, 147.e1–147.e8 (2007).
Fitzgibbons, S. C. et al. Mortality of necrotizing enterocolitis expressed by birth weight categories. J. Pedia. Surg. 44, 1072–1076 (2009).
Hintz, S. R. Neurodevelopmental and growth outcomes of extremely low birth weight infants after necrotizing enterocolitis. Pediatrics 115, 696–703 (2005).
Park, H. W., Lee, N. M., Kim, J. H., Kim, K. S. & Kim, S. N. Parenteral fish oil-containing lipid emulsions may reverse parenteral nutrition-associated cholestasis in neonates: a systematic review and meta-analysis. J. Nutr. 145, 277–283 (2015).
Teitelbaum, D. H. & Tracy, T. Parenteral nutrition-associated cholestasis. Semin Pedia. Surg. 10, 72–80 (2001).
Pammi, M. et al. Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: a systematic review and meta-analysis. Microbiome 5, 1–15 (2017).
Korpela, K. et al. Intestinal microbiota signatures associated with histological liver steatosis in pediatric-onset intestinal failure. JPEN 41, 238–248 (2017).
Harris, P. A. et al. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inf. 42, 377–381 (2009).
Edgar, R. C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nature 10, 996–998 (2013).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2012).
Methé, B. A. et al. A framework for human microbiome research. Nature 486, 215–221 (2012).
Aagaard, K. et al. The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB J. 27, 1012–1022 (2013).
Harvey, R. B. et al. Qualitative and quantitative comparison of gut bacterial colonization in enterally and parenterally fed neonatal pigs. Curr. Issues Intest Microbiol 7, 61–64 (2006).
Huang, Y., Guo, F., Li, Y., Wang, J. & Li, J. Fecal microbiota signatures of adult patients with different types of short bowel syndrome. J. Gastroenterol. Hepatol. 32, 1949–1957 (2017).
Lilja, H. E., Wefer, H., Nyström, N., Finkel, Y. & Engstrand, L. Intestinal dysbiosis in children with short bowel syndrome is associated with impaired outcome. Microbiome 3, 1–6 (2015).
Shiga, H. et al. Changes of faecal microbiota in patients with Crohn’s disease treated with an elemental diet and total parenteral nutrition. Dig. Liver Dis. 44, 736–742 (2012).
Parm, Ü., Metsvaht, T., Ilmoja, M.-L. & Lutsar, I. Gut colonization by aerobic microorganisms is associated with route and type of nutrition in premature neonates. Nutr. Res. 35, 496–503 (2015).
Chu, D. M. et al. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med 23, 314–326 (2017).
Jost, T., Lacroix, C., Braegger, C. P. & Chassard, C. New insights in gut microbiota establishment in healthy breast fed neonates. PLoS ONE 7, e44595 (2012).
Mazmanian, S. K., Round, J. L. & Kasper, D. L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625 (2008).
Troy, E. B. & Kasper, D. L. Beneficial effects of Bacteroides fragilis polysaccharides on the immune system. Front Biosci. 15, 25–34 (2010).
Ganguli, K. et al. Probiotics prevent necrotizing enterocolitis by modulating enterocyte genes that regulate innate immune-mediated inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 304, G132–G141 (2013).
Fukuda, S. et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469, 543–547 (2011).
Belzer, C. & de Vos, W. M. Microbes inside—from diversity to function: the case of Akkermansia. ISME J. 6, 1449–1458 (2012).
Shin, N.-R. et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63, 727–735 (2014).
Seregin, S. S. et al. NLRP6 protects Il10−/− mice from colitis by limiting colonization of Akkermansia muciniphila. Cell Rep. 19, 733–745 (2017).
Durand, L. et al. Microbial diversity associated with the hydrothermal shrimp Rimicaris exoculatagut and occurrence of a resident microbial community. FEMS Microbiol Ecol. 71, 291–303 (2010).
Bäckhed, F. et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 690–703 (2015).
Gupta, R. W. et al. Histamine-2 receptor blockers alter the fecal microbiota in premature infants. J. Pedia. Gastroenterol. Nutr. 56, 397–400 (2013).
Wang, Y. et al. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 3, 944–954 (2009).
Oddie, S. J., Young, L. & McGuire, W. Slow advancement of enteral feed volumes to prevent necrotising enterocolitis in very low birth weight infants. Cochrane Database Syst. Rev. 114, 1597–37 (2017).
Rozé, J. C. et al. Nutritional strategies and gut microbiota composition as risk factors for necrotizing enterocolitis in very-preterm infants. Am. J. Clin. Nutr. 106, 821–830 (2017).
Wang, P. et al. Alterations in intestinal microbiota relate to intestinal failure-associated liver disease and central line infections. J. Pedia. Surg. 52, 1318–1326 (2017).
Wildhaber, B. E., Yang, H., Spencer, A. U., Drongowski, R. A. & Teitelbaum, D. H. Lack of enteral nutrition—effects on the intestinal immune system. J. Surg. Res. 123, 8–16 (2005).
Ralls, M. W., Demehri, F. R., Feng, Y., Woods Ignatoski, K. M. & Teitelbaum, D. H. Enteral nutrient deprivation in patients leads to a loss of intestinal epithelial barrier function. Surgery 157, 732–742 (2015).
Acknowledgements
We would like to thank the Children’s Hospital of Wisconsin Pediatric Translational Research Unit for assistance with sample collection and processing, and Alec Monaghan, BS, for his assistance with data collection. These individuals have no conflicts of interest to disclose. Research reported in this publication was supported by the National Institutes of Health (K23 DK109071 [to T.H.N.-R.], R35 GM122503 and R01 GM099526 [to N.H.S.]), and the Gerber Foundation [to T.H.N.-R.]. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the various funding agencies.
Author information
Authors and Affiliations
Contributions
Each author has met the requirements for authorship. Substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data were made by A.F.D., A.P., V.L., K.C.G., P.M.S., N.H.S. and T.H.N.-R. The first draft of the manuscript was written by T.H.N.-R., and all authors contributed to revising the manuscript critically for important intellectual content. Each author has seen and approved the submission of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Dahlgren, A.F., Pan, A., Lam, V. et al. Longitudinal changes in the gut microbiome of infants on total parenteral nutrition. Pediatr Res 86, 107–114 (2019). https://doi.org/10.1038/s41390-019-0391-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-019-0391-y
This article is cited by
-
Levofloxacin prophylaxis and parenteral nutrition have a detrimental effect on intestinal microbial networks in pediatric patients undergoing HSCT
Communications Biology (2023)
-
Gut microbiota disruption during sepsis and the influence of innate metabolites on sepsis prognosis
International Microbiology (2023)
-
The central and biodynamic role of gut microbiota in critically ill patients
Critical Care (2022)
-
Probiotic supplementation in neonates with congenital gastrointestinal surgical conditions: a pilot randomised controlled trial
Pediatric Research (2022)
-
Neonatal intensive care unit (NICU) exposures exert a sustained influence on the progression of gut microbiota and metabolome in the first year of life
Scientific Reports (2021)